1E5D
 
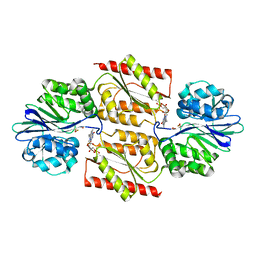 | | RUBREDOXIN OXYGEN:OXIDOREDUCTASE (ROO) FROM ANAEROBE DESULFOVIBRIO GIGAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, MU-OXO-DIIRON, OXYGEN MOLECULE, ... | | Authors: | Frazao, C, Silva, G, Gomes, C.M, Matias, P, Coelho, R, Sieker, L, Macedo, S, Liu, M.Y, Oliveira, S, Teixeira, M, Xavier, A.V, Rodrigues-Pousada, C, Carrondo, M.A, Le Gall, J. | | Deposit date: | 2000-07-24 | | Release date: | 2000-11-17 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Nat.Struct.Biol., 7, 2000
|
|
1H1D
 
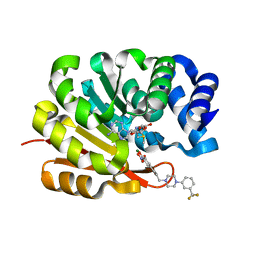 | | Catechol O-Methyltransferase | | Descriptor: | 1-(3,4,DIHYDROXY-5-NITROPHENYL)-3-{4-[3-(TRIFLUOROMETHYL) PHENYL] PIPERAZIN-1-YL}PROPAN-1-ONE, CATECHOL-O-METHYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Archer, M, Rodrigues, M.L, Matias, P.M, Bonifacio, M.J, Learmonth, D.A, Soares-da-Silva, P, Carrondo, M.A. | | Deposit date: | 2002-07-12 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetics and Crystal Structure of Catechol-O-Methyltransferase Complex with Co-Substrate and a Novel Inhibitor with Potential Therapeutic Application
Mol.Pharmacol., 62, 2002
|
|
1OF0
 
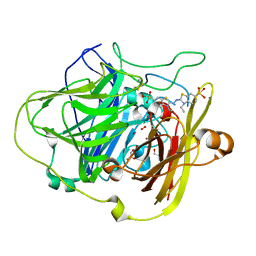 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS COTA AFTER 1H SOAKING WITH ABTS | | Descriptor: | 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (I) ION, CU-O LINKAGE, ... | | Authors: | Enguita, F.J, Marcal, D, Grenha, R, Martins, L.O, Henriques, A.O, Carrondo, M.A. | | Deposit date: | 2003-04-03 | | Release date: | 2004-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Characterization of a Bacterial Laccase Reaction Cycle
To be Published
|
|
1OFY
 
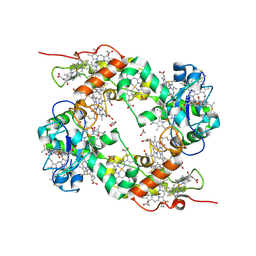 | | three dimensional structure of the reduced form of nine-heme cytochrome c at ph 7.5 | | Descriptor: | ACETATE ION, GLYCEROL, HEME C, ... | | Authors: | Bento, I, Teixeira, V.H, Baptista, A.M, Soares, C.M, Matias, P.M, Carrondo, M.A. | | Deposit date: | 2003-04-22 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-Bohr and Other Cooperativity Effects in the Nine-Heme Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies
J.Biol.Chem., 278, 2003
|
|
1OFW
 
 | | Three dimensional structure of the oxidized form of nine heme cytochrome c at PH 7.5 | | Descriptor: | ACETATE ION, GLYCEROL, HEME C, ... | | Authors: | Bento, I, Teixeira, V.H, Baptista, A.M, Soares, C.M, Matias, P.M, Carrondo, M.A. | | Deposit date: | 2003-04-22 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-Bohr and Other Cooperativity Effects in the Nine-Heme Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774: Crystallographic and Modeling Studies
J.Biol.Chem., 278, 2003
|
|
1OAE
 
 | | Crystal structure of the reduced form of cytochrome c" from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C", GLYCEROL, HEME C, ... | | Authors: | Enguita, F.J, Grenha, R, Santos, H, Carrondo, M.A. | | Deposit date: | 2003-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
4D02
 
 | | The crystallographic structure of Flavorubredoxin from Escherichia coli | | Descriptor: | ANAEROBIC NITRIC OXIDE REDUCTASE FLAVORUBREDOXIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Romao, C.V, Vicente, J.B, Bandeiras, T, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2014-04-23 | | Release date: | 2014-09-03 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structure of Escherichia Coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
4AJB
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with Isocitrate, magnesium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJS
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate, magnesium(II), Adenosine 2',5'-biphosphate and ribosylnicotinamide-5'-phosphate | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJR
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with alpha-ketoglutarate, magnesium(II) and NADPH - The product complex | | Descriptor: | 2-OXOGLUTARIC ACID, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ISOCITRATE DEHYDROGENASE [NADP], ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4C7U
 
 | | Crystal structure of manganese superoxide dismutase from Arabidopsis thaliana | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE [MN] 1, MITOCHONDRIAL | | Authors: | Marques, A, Santos, S.P, Rosa, M, Carrondo, M.A, Abreu, I.A, Romao, C.V, Frazao, C. | | Deposit date: | 2013-09-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of the Arabidopsis Thaliana Manganese Superoxide Dismutase at 1.95 A Resolution
To be Published
|
|
4COT
 
 | | The importance of the Abn2 calcium cluster in the endo-1,5- arabinanase activity from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EXTRACELLULAR ENDO-ALPHA-(1->5)-L-ARABINANASE 2, NICKEL (II) ION | | Authors: | McVey, C.E, Ferreira, M.J, Correia, B, Lahiri, S, deSanctis, D, Carrondo, M.A, Lindley, P.F, de Sa-Nogueira, I, Soares, C.M, Bento, I. | | Deposit date: | 2014-01-31 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Importance of the Abn2 Calcium Cluster in the Endo-1,5-Arabinanase Activity from Bacillus Subtilis.
J.Biol.Inorg.Chem., 19, 2014
|
|
4AJ3
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and NADP - The pseudo-Michaelis complex | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, NADP ISOCITRATE DEHYDROGENASE, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4AJA
 
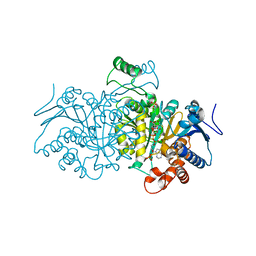 | | 3D structure of E. coli Isocitrate Dehydrogenase in complex with Isocitrate, calcium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
4BLG
 
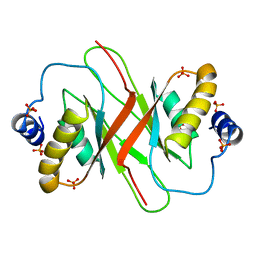 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
1E3K
 
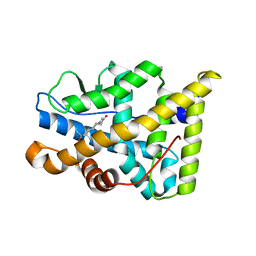 | | Human Progesteron Receptor Ligand Binding Domain in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, PROGESTERONE RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1E3D
 
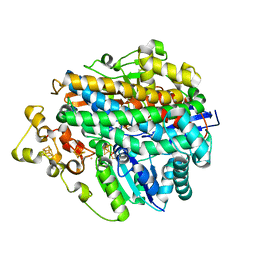 | | [NiFe] Hydrogenase from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), FE3-S4 CLUSTER, ... | | Authors: | Matias, P.M, Soares, C.M, Saraiva, L.M, Coelho, R, Morais, J, Le Gall, J, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | [Nife] Hydrogenase from Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of its Interaction with the Tetrahaem Cytochrome C3.
J.Biol.Inorg.Chem., 6, 2001
|
|
1E3G
 
 | | Human Androgen Receptor Ligand Binding in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, ANDROGEN RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Ruff, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1GNL
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans X-ray structure at 1.25A resolution using synchrotron radiation at a wavelength of 0.933A | | Descriptor: | ACETATE ION, HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GN9
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans ATCC 27774 X-ray structure at 2.6A resolution using synchrotron radiation at a wavelength of 1.722A | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-04 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
1GS4
 
 | | Structural basis for the glucocorticoid response in a mutant human androgen receptor (ARccr) derived from an androgen-independent prostate cancer | | Descriptor: | 9ALPHA-FLUOROCORTISOL, ANDROGEN RECEPTOR, PHOSPHATE ION | | Authors: | Matias, P.M, Carrondo, M.A, Coelho, R, Thomaz, M, Zhao, X.-Y, Wegg, A, Crusius, K, Egner, U, Donner, P. | | Deposit date: | 2001-12-27 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Glucocorticoid Response in a Mutant Human Androgen Receptor (Ar(Ccr)) Derived from an Androgen-Independent Prostate Cancer
J.Med.Chem., 45, 2002
|
|
1GU2
 
 | | Crystal structure of oxidized cytochrome c'' from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Enguita, F.J, Pohl, E, Rodrigues, A, Santos, H, Carrondo, M.A. | | Deposit date: | 2002-01-22 | | Release date: | 2003-01-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
1GNT
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris. X-ray structure at 1.25A resolution using synchrotron radiation. | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hybrid cluster proteins (HCPs) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at 1.25 A resolution using synchrotron radiation.
J. Biol. Inorg. Chem., 7, 2002
|
|
1W9M
 
 | | AS-isolated hybrid cluster protein from Desulfovibrio vulgaris X-ray structure at 1.35A resolution using iron anomalous signal | | Descriptor: | FE-S-O HYBRID CLUSTER, HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Coelho, D, Romao, C.V, Teixeira, M, Lindley, P.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-02-04 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Relationships in the Hybrid Cluster Protein Family:Structure of the Anaerobically Purified Hybrid Cluster Protein from Desulfovibrio Vulgaris at 1.35 A Resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5A76
 
 | |
