4N6N
 
 | | Crystal structure of oxidized legumain in complex with cystatin E/M | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cystatin-M, IODIDE ION, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2013-10-14 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure and mechanism of an aspartimide-dependent Peptide ligase in human legumain.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4N1K
 
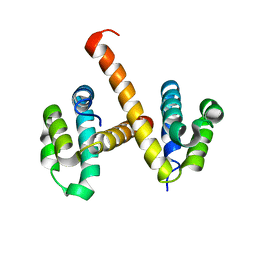 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N6L
 
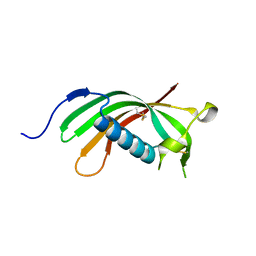 | | Crystal structure of human cystatin E/M | | Descriptor: | Cystatin-M | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2013-10-14 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structure and mechanism of an aspartimide-dependent Peptide ligase in human legumain.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4NFF
 
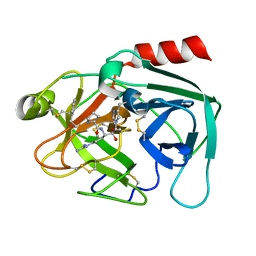 | | Human kallikrein-related peptidase 2 in complex with PPACK | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Kallikrein-2 | | Authors: | Skala, W, Brandstetter, H, Magdolen, V, Goettig, P. | | Deposit date: | 2013-10-31 | | Release date: | 2014-10-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity
J.Biol.Chem., 289, 2014
|
|
4NFE
 
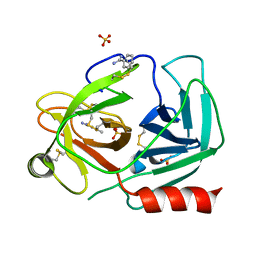 | | Human kallikrein-related peptidase 2 in complex with benzamidine | | Descriptor: | BENZAMIDINE, Kallikrein-2, SULFATE ION | | Authors: | Skala, W, Brandstetter, H, Magdolen, V, Goettig, P. | | Deposit date: | 2013-10-31 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity
J.Biol.Chem., 289, 2014
|
|
1JH1
 
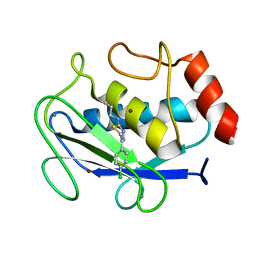 | | Crystal Structure of MMP-8 complexed with a 6H-1,3,4-thiadiazine derived inhibitor | | Descriptor: | BUT-3-ENYL-[5-(4-CHLORO-PHENYL)-3,6-DIHYDRO-[1,3,4]THIADIAZIN-2-YLIDENE]-AMINE, CALCIUM ION, Matrix Metalloproteinase 8, ... | | Authors: | Schroder, J, Henke, A, Wenzel, H, Brandstetter, H, Stammler, H.G, Stammler, A, Pfeiffer, W.D, Tschesche, H. | | Deposit date: | 2001-06-27 | | Release date: | 2001-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based design and synthesis of potent matrix metalloproteinase inhibitors derived from a 6H-1,3,4-thiadiazine scaffold.
J.Med.Chem., 44, 2001
|
|
1J2Q
 
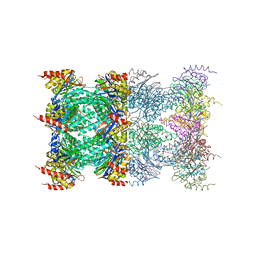 | | 20S proteasome in complex with calpain-Inhibitor I from archaeoglobus fulgidus | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID [1-(1-FORMYL-PENTYLCARBAMOYL)-3-METHYL-BUTYL]-AMIDE, Proteasome alpha subunit, Proteasome beta subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
1J2P
 
 | | alpha-ring from the proteasome from archaeoglobus fulgidus | | Descriptor: | Proteasome alpha subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
1ITV
 
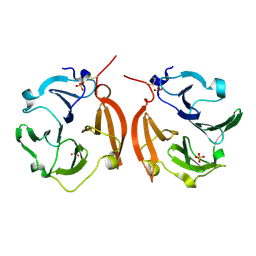 | | Dimeric form of the haemopexin domain of MMP9 | | Descriptor: | MMP9, SULFATE ION | | Authors: | Cha, H, Kopetzki, E, Huber, R, Lanzendoerfer, M, Brandstetter, H. | | Deposit date: | 2002-02-11 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the adaptive molecular recognition by MMP9.
J.Mol.Biol., 320, 2002
|
|
1S0P
 
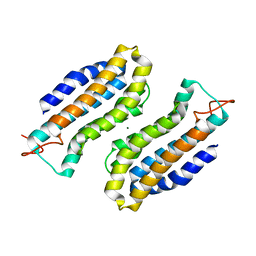 | | Structure of the N-Terminal Domain of the Adenylyl Cyclase-Associated Protein (CAP) from Dictyostelium discoideum. | | Descriptor: | Adenylyl cyclase-associated protein, MAGNESIUM ION | | Authors: | Ksiazek, D, Brandstetter, H, Israel, L, Bourenkov, G.P, Katchalova, G, Janssen, K.P, Bartunik, H.D, Noegel, A.A, Schleicher, M, Holak, T.A. | | Deposit date: | 2004-01-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL
CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Structure, 11, 2003
|
|
1RFN
 
 | | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | | Descriptor: | CALCIUM ION, P-AMINO BENZAMIDINE, PROTEIN (COAGULATION FACTOR IX), ... | | Authors: | Hopfner, K.-P, Lang, A, Karcher, A, Sichler, K, Kopetzki, E, Brandstetter, H, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1999-04-19 | | Release date: | 1999-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Structure Fold.Des., 7, 1999
|
|
4BQD
 
 | | KD1 of human TFPI in complex with a synthetic peptide | | Descriptor: | GLYCEROL, PEPTIDE, TISSUE FACTOR PATHWAY INHIBITOR (LIPOPROTEIN-ASSOCIATED COAGULATION INHIBITOR) VARIANT | | Authors: | Griessner, A, Brandstetter, H. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Small Peptides Blocking Inhibition of Factor Xa and Tissue Factor-Factor Viia by Tissue Factor Pathway Inhibitor (Tfpi)
J.Biol.Chem., 289, 2014
|
|
3ZFP
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with internal His-Tag | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFT
 
 | | Crystal structure of product-like, processed N-terminal protease Npro at pH 3 | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | Descriptor: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFU
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P with sulphate | | Descriptor: | MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO, SULFATE ION | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFR
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with iridium | | Descriptor: | HYDROXIDE ION, IRIDIUM (III) ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFQ
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with mercury | | Descriptor: | MERCURY (II) ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFN
 
 | | Crystal structure of product-like, processed N-terminal protease Npro | | Descriptor: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
4A83
 
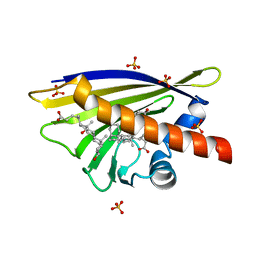 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in complex with deoxycholate. | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
4A8G
 
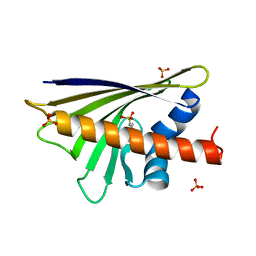 | |
4ARF
 
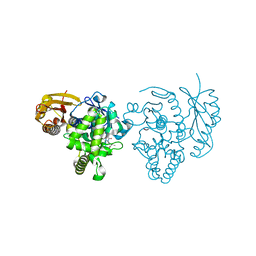 | | CRYSTAL STRUCTURE OF THE PEPTIDASE DOMAIN OF COLLAGENASE H FROM CLOSTRIDIUM HISTOLYTICUM IN COMPLEX WITH THE PEPTIDIC INHIBITOR ISOAMYLPHOSPHONYL-GLY-PRO-ALA AT 1.77 ANGSTROM RESOLUTION. | | Descriptor: | CALCIUM ION, COLH PROTEIN, ISOAMYLPHOSPHONYL-GLY-PRO-ALA, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-23 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
4A8V
 
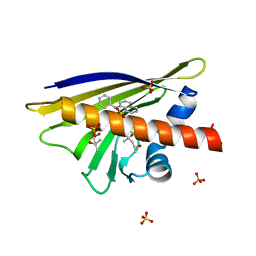 | | Crystal Structure of Birch Pollen Allergen Bet v 1 isoform j in complex with 8-Anilinonaphthalene-1-sulfonate (ANS) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
4A8U
 
 | | Crystal Structure of native Birch Pollen Allergen Bet v 1 isoform j | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
1MT3
 
 | | Crystal Structure of the Tricorn Interacting Factor Selenomethionine-F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
