7NFC
 
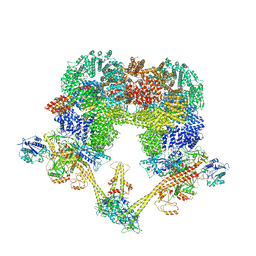 | | Cryo-EM structure of NHEJ super-complex (dimer) | | Descriptor: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-18 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFE
 
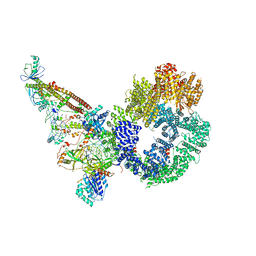 | | Cryo-EM structure of NHEJ super-complex (monomer) | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-06 | | Release date: | 2021-08-18 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
1E0O
 
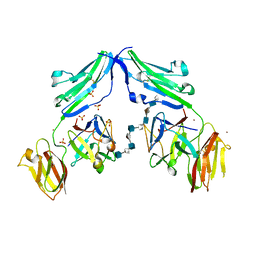 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
7OR2
 
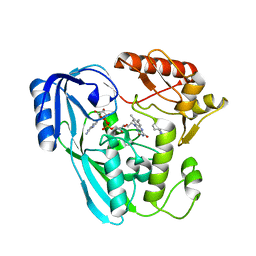 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OSQ
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7ORZ
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-06 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OTM
 
 | | Cryo-EM structure of DNA-PKcs in complex with NU7441 | | Descriptor: | 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
7OTP
 
 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
1GMN
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1GMO
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1N0W
 
 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
3H4T
 
 | | Chimeric Glycosyltransferase for the generation of novel natural products - GtfAH1 in complex with UDP-2F-Glc | | Descriptor: | Glycosyltransferase GtfA, Glycosyltransferase, PHOSPHATE ION, ... | | Authors: | Dias, M.V.B, Truman, A.W, Wu, S, Blundell, T.L, Huang, F, Spencer, J.B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Chimeric glycosyltransferases for the generation of hybrid glycopeptides
Chem.Biol., 16, 2009
|
|
3H4I
 
 | | Chimeric Glycosyltransferase for the generation of novel natural products | | Descriptor: | Glycosyltransferase GtfA, Glycosyltransferase, SULFATE ION, ... | | Authors: | Dias, M.V.B, Truman, A.W, Wu, S, Blundell, T.L, Huang, F, Spencer, J.B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chimeric glycosyltransferases for the generation of hybrid glycopeptides
Chem.Biol., 16, 2009
|
|
3HWO
 
 | |
3ISJ
 
 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-methoxy-N-(methylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 5-methoxy-N-(methylsulfonyl)-1H-indole-2-carboxamide, ETHANOL, GLYCEROL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IMG
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.8 Ang resolution in a ternary complex with fragment compounds 5-methoxyindole and 1-benzofuran-2-carboxylic acid | | Descriptor: | 1-benzofuran-2-carboxylic acid, 5-methoxy-1H-indole, ETHANOL, ... | | Authors: | Ciulli, A, Hung, A.W, Silvestre, H.L, Wen, S, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1PDA
 
 | | STRUCTURE OF PORPHOBILINOGEN DEAMINASE REVEALS A FLEXIBLE MULTIDOMAIN POLYMERASE WITH A SINGLE CATALYTIC SITE | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, PORPHOBILINOGEN DEAMINASE | | Authors: | Louie, G.V, Brownlie, P.D, Lambert, R, Cooper, J.B, Blundell, T.L, Wood, S.P, Warren, M.J, Woodcock, S.C, Jordan, P.M. | | Deposit date: | 1992-11-17 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site.
Nature, 359, 1992
|
|
3IUB
 
 | | Crystal structure of pantothenate synthetase from Mycobacterium tuberculosis in complex with 5-Methoxy-N-(5-methylpyridin-2-ylsulfonyl)-1H-indole-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-methoxy-N-[(5-methylpyridin-2-yl)sulfonyl]-1H-indole-2-carboxamide, ETHANOL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IUE
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(5-methoxy-2-(5-Methylpyridin-2-ylsulfonylcarbamoyl)-1H-indol-1-yl) acetic acid | | Descriptor: | (5-methoxy-2-{[(5-methylpyridin-2-yl)sulfonyl]carbamoyl}-1H-indol-1-yl)acetic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L, Wen, S, Hung, A.W, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IMC
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.6 Ang resolution in complex with fragment compound 5-methoxyindole, sulfate and glycerol | | Descriptor: | 5-methoxy-1H-indole, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Hung, A.W, Silvestre, H.L, Wen, S, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IVC
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(2-((benzofuran-2-ylmethoxy)carbonyl)-5-methoxy-1H-indol-1-yl)acetic acid | | Descriptor: | ETHANOL, GLYCEROL, Pantothenate synthetase, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3CMS
 
 | | ENGINEERING ENZYME SUB-SITE SPECIFICITY: PREPARATION, KINETIC CHARACTERIZATION AND X-RAY ANALYSIS AT 2.0-ANGSTROMS RESOLUTION OF VAL111PHE SITE-MUTATED CALF CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Shearer, A, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1990-02-26 | | Release date: | 1992-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering enzyme subsite specificity: preparation, kinetic characterization, and X-ray analysis at 2.0-A resolution of Val111Phe site-mutated calf chymosin.
Biochemistry, 29, 1990
|
|
3IVX
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(2-(benzofuran-2-ylsulfonylcarbamoyl)-5-methoxy-1H-indol-1-yl)acetic acid | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, GLYCEROL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-09-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IVG
 
 | | Crystal structure of pantothenate synthetase in complex with 2-(2-((benzofuran-2-sulfonamido)methyl)-5-methoxy-1H-indol-1-yl)acetic acid | | Descriptor: | (2-{[(1-benzofuran-2-ylsulfonyl)amino]methyl}-5-methoxy-1H-indol-1-yl)acetic acid, 1,2-ETHANEDIOL, ETHANOL, ... | | Authors: | Silvestre, H.L, Hung, A.W, Wen, S, Ciulli, A, Blundell, T.L, Abell, C. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
