8FS1
 
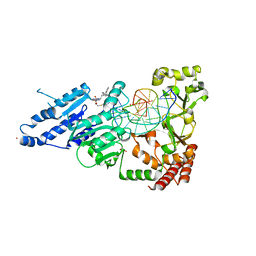 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
2JUT
 
 | |
2JSY
 
 | | Solution structure of Tpx in the oxidized state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2JUR
 
 | |
2JUQ
 
 | |
5IKL
 
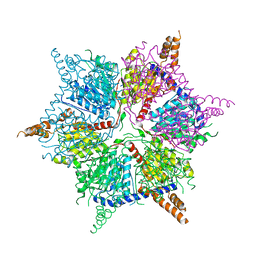 | |
8IYJ
 
 | | Cryo-EM structure of the 48-nm repeat doublet microtubule from mouse sperm | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhou, L.N, Gui, M, Wu, J.P. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of sperm flagellar doublet microtubules expand the genetic spectrum of male infertility.
Cell, 186, 2023
|
|
7STI
 
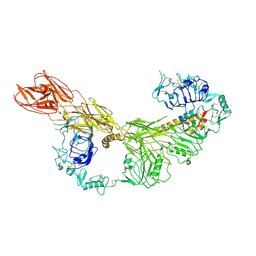 | |
7STJ
 
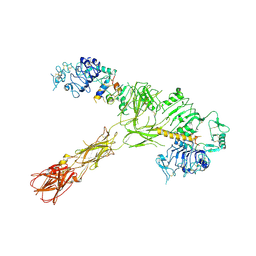 | |
7STK
 
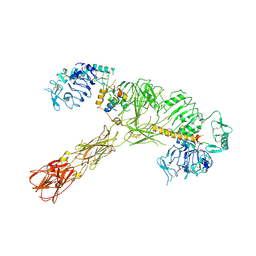 | |
7STH
 
 | |
7T5S
 
 | | P. aeruginosa LpxA in complex with ligand H16 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, N~2~-(cyclohexylacetyl)-N-1H-tetrazol-5-yl-L-alaninamide | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5R
 
 | | P. aeruginosa LpxA in complex with ligand H7 | | Descriptor: | 3-bromo-N-[3-(1H-tetrazol-5-yl)phenyl]-1H-indole-5-carboxamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5X
 
 | | P. aeruginosa LpxA in complex with ligand L6 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, Nalpha-(tert-butoxycarbonyl)-N-1H-tetrazol-5-yl-D-tryptophanamide | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5Z
 
 | | P. aeruginosa LpxA in complex with ligand L8 | | Descriptor: | (4S)-N-(1H-tetrazol-5-yl)-2-[3-(trifluoromethyl)benzene-1-sulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T60
 
 | | P. aeruginosa LpxA in complex with ligand L13 | | Descriptor: | (3S)-3-(5,5-dimethyl-2-oxo-1,3-oxazolidin-3-yl)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T61
 
 | | P. aeruginosa LpxA in complex with ligand L15 | | Descriptor: | (3S)-3-({[(Z)-phenylmethylidene]carbamoyl}amino)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, ACETATE ION, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
5YAD
 
 | | Crystal structure of Marf1 Lotus domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1, SULFATE ION | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YAA
 
 | | Crystal structure of Marf1 NYN domain from Mus musculus | | Descriptor: | GLYCEROL, Meiosis regulator and mRNA stability factor 1 | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2017-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7TOB
 
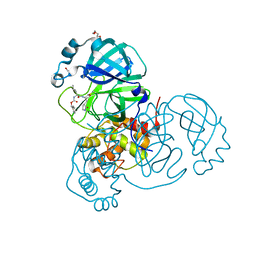 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M.D, Wang, J, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
7F6V
 
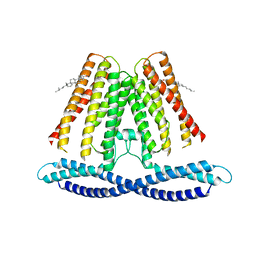 | | Cryo-EM structure of the human TACAN channel in a closed state | | Descriptor: | CHOLESTEROL, Ion channel TACAN | | Authors: | Chen, X.Z, Wang, Y.J, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2021-06-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structure of the human TACAN in a closed state.
Cell Rep, 38, 2022
|
|
8DWB
 
 | |
8DMK
 
 | |
8E2B
 
 | | N-terminal domain of S. aureus GpsB | | Descriptor: | Cell cycle protein GpsB, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Elife, 13, 2024
|
|
8E2C
 
 | |
