1L3I
 
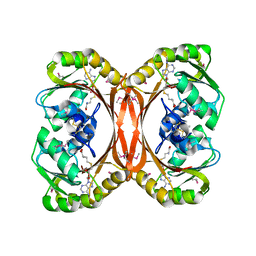 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, ADOHCY BINARY COMPLEX | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1NPD
 
 | | X-RAY STRUCTURE OF SHIKIMATE DEHYDROGENASE COMPLEXED WITH NAD+ FROM E.COLI (YDIB) NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER24 | | Descriptor: | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Kuzin, A.P, Lee, I, Rost, B, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3-A crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase
J.Biol.Chem., 278, 2003
|
|
1L2T
 
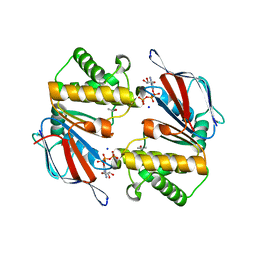 | | Dimeric Structure of MJ0796, a Bacterial ABC Transporter Cassette | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hypothetical ABC transporter ATP-binding protein MJ0796, ISOPROPYL ALCOHOL, ... | | Authors: | Smith, P.C, Karpowich, N, Rosen, J, Hunt, J.F. | | Deposit date: | 2002-02-24 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer.
Mol.Cell, 10, 2002
|
|
1L3B
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP W/ LONG CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1OY1
 
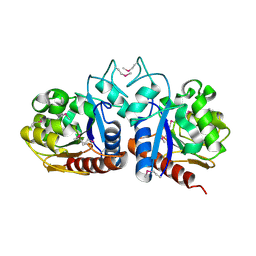 | | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105 | | Descriptor: | PUTATIVE sigma cross-reacting protein 27A | | Authors: | Benach, J, Edstrom, W, Ma, L.C, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105
To be Published
|
|
1NS5
 
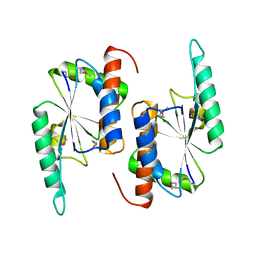 | | X-RAY STRUCTURE OF YBEA FROM E.COLI. NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER45 | | Descriptor: | Hypothetical protein ybeA | | Authors: | Benach, J, Shen, J, Rost, B, Xiao, R, Acton, T, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of YBEA from E. coli
To be Published
|
|
1QYI
 
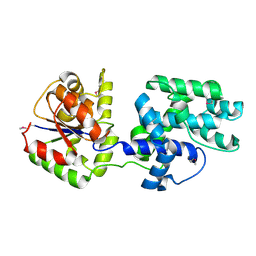 | | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25. | | Descriptor: | hypothetical protein | | Authors: | Kuzin, A.P, Edstrom, W, Ma, L.C, Shin, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-10 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25.
To be Published
|
|
1KXZ
 
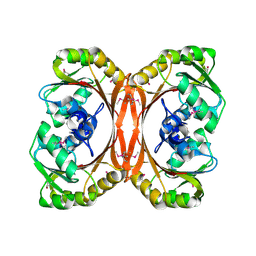 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3C
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP WITH SHORT CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of Mt0146/Cbit Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1NY1
 
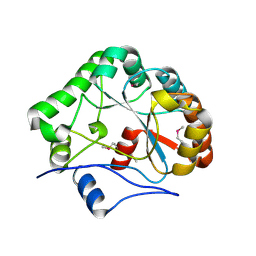 | | CRYSTAL STRUCTURE OF B. SUBTILIS POLYSACCHARIDE DEACETYLASE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR127. | | Descriptor: | Probable polysaccharide deacetylase pdaA | | Authors: | Forouhar, F, Edstrom, W, Khan, J, Ma, L, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Polysaccharide Deacetylase (PDAA_BACSU) from B. Subtilis (Pdaa_Bacsu) Northeast Structural Genomics Research Consortium (Nesg) Target Sr127
To be Published
|
|
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
1MZH
 
 | | QR15, an Aldolase | | Descriptor: | Deoxyribose-phosphate aldolase, PHOSPHATE ION | | Authors: | Tan, A.Y, Smith, P.C, Shen, J, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aquifex Aeolicus Aldolase,
Northeast Structural Genomics Consortium Target
QR15
To be Published
|
|
1JOD
 
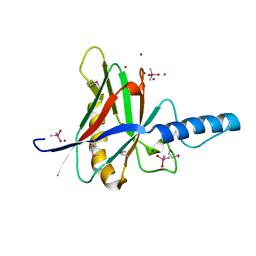 | |
1N4A
 
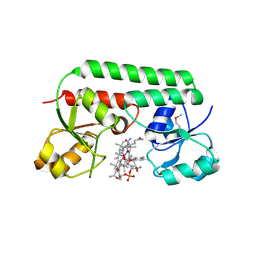 | | The Ligand Bound Structure of E.coli BtuF, the Periplasmic Binding Protein for Vitamin B12 | | Descriptor: | CYANOCOBALAMIN, Vitamin B12 transport protein btuF | | Authors: | Karpowich, N.K, Smith, P.C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-30 | | Release date: | 2003-03-11 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding.
J.Biol.Chem., 278, 2003
|
|
1MW7
 
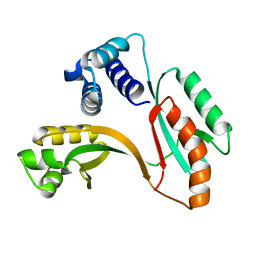 | | X-RAY STRUCTURE OF Y162_HELPY NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET PR6 | | Descriptor: | HYPOTHETICAL PROTEIN HP0162 | | Authors: | Kuzin, A, Shen, J, Keller, J.P, Xiao, R, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-09-27 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE OF Y162_HELPY NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET PR6
To be published
|
|
1N4D
 
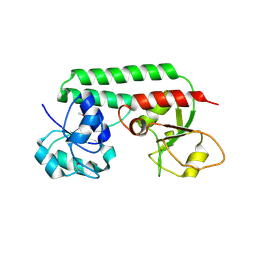 | |
1RTW
 
 | | X-ray Structure of PF1337, a TenA Homologue from Pyrococcus furiosus. Northeast Structural Genomics Research Consortium (Nesg) Target PFR34 | | Descriptor: | (4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, transcriptional activator, ... | | Authors: | Benach, J, Edstrom, W.C, Lee, I, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The 2.35 A structure of the TenA homolog from Pyrococcus furiosus supports an enzymatic function in thiamine metabolism.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4E88
 
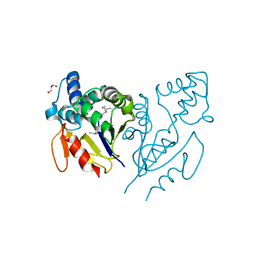 | | CRYSTAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, GLYCEROL | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Xiao, X, Sahdev, S, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
4EVW
 
 | | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9. Northeast Structural Genomics Consortium (NESG) Target VcR193. | | Descriptor: | MAGNESIUM ION, Nucleoside-diphosphate-sugar pyrophosphorylase | | Authors: | Vorobiev, S, Neely, H, Su, M, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9.
To be Published
|
|
4ESS
 
 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4E0A
 
 | | Crystal Structure of the mutant F44R BH1408 protein from Bacillus halodurans, Northeast Structural Genomics Consortium (NESG) Target BhR182 | | Descriptor: | ACETIC ACID, BH1408 protein, GLYCEROL, ... | | Authors: | Kuzin, A, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-02 | | Release date: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Northeast Structural Genomics Consortium Target BhR182
To be Published
|
|
4DMB
 
 | | X-ray structure of human hepatitus C virus NS5A-transactivated protein 2 at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HR6723 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HD domain-containing protein 2, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-02-07 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR6723
To be Published
|
|
4DIU
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94 | | Descriptor: | Engineered Protein PF00326 | | Authors: | Seetharaman, J, Lew, S, Wang, D, Kohan, E, Patel, D, Whitehead, T, Fleishman, S, Ciccosanti, C, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94
To be Published
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
