6DGF
 
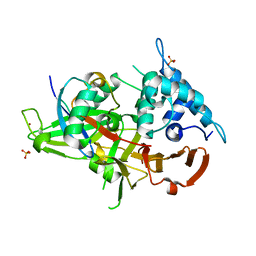 | | Ubiquitin Variant bound to USP2 | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 2, ... | | Authors: | Manczyk, N, Sicheri, F. | | Deposit date: | 2018-05-17 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Yeast Two-Hybrid Analysis for Ubiquitin Variant Inhibitors of Human Deubiquitinases.
J. Mol. Biol., 431, 2019
|
|
6DMR
 
 | |
6DMU
 
 | | PI(4,5)P2 bound full-length rbTRPV5 | | Descriptor: | Transient receptor potential cation channel subfamily V member 5, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hughes, T.E.T, Pumroy, R.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights on TRPV5 gating by endogenous modulators.
Nat Commun, 9, 2018
|
|
6DMW
 
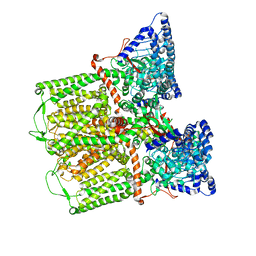 | |
7CSQ
 
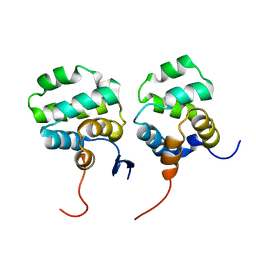 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | Descriptor: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
5JFG
 
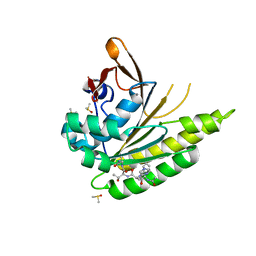 | | Structure of humanised RadA-mutant humRadA22F in complex with peptide FHTA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Humanisation of RadA from Pyrococcus furiosus
To Be Published
|
|
3KVL
 
 | |
3KVJ
 
 | |
4K3D
 
 | | Crystal structure of bovine antibody BLV1H12 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K3E
 
 | | Crystal structure of bovine antibody BLV5B8 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
5D6X
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5D6Y
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
3N2X
 
 | | Crystal structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 in complex with pyruvate | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-05-19 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
5EAJ
 
 | | Crystal structure of DHFR in 0% Isopropanol | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cuneo, M.J, Agarwal, P.K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Modulating Enzyme Activity by Altering Protein Dynamics with Solvent.
Biochemistry, 57, 2018
|
|
5D6W
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5EFM
 
 | | Beclin 1 Flexible-helical Domian (FHD) (141-171) | | Descriptor: | Beclin-1, SULFATE ION | | Authors: | Sinha, S, Mei, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational Flexibility Enables the Function of a BECN1 Region Essential for Starvation-Mediated Autophagy.
Biochemistry, 55, 2016
|
|
3KVM
 
 | |
3KVK
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with amino-benzoic acid inhibitor 641 at 2.05A resolution | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 2-{[(3,5-dichlorophenyl)carbamoyl]amino}benzoic acid, Dihydroorotate dehydrogenase, ... | | Authors: | McLean, L, Zhang, Y. | | Deposit date: | 2009-11-30 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of novel inhibitors for DHODH via virtual screening and X-ray crystallographic structures.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5E3H
 
 | | Structural Basis for RNA Recognition and Activation of RIG-I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Jiang, F, Miller, M.T, Marcotrigiano, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of RNA recognition and activation by innate immune receptor RIG-I.
Nature, 479, 2011
|
|
3NEV
 
 | | Crystal structure of YagE, a prophage protein from E. coli K12 in complex with KDGal | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-LYXO-HEXONIC ACID, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
2ZEQ
 
 | | Crystal structure of ubiquitin-like domain of murine Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin | | Authors: | Tomoo, K. | | Deposit date: | 2007-12-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkin
Biochim.Biophys.Acta, 1784, 2008
|
|
2G8Q
 
 | |
2G8R
 
 | | The crystal structure of the RNase A- 3-N-piperidine-4-carboxyl-3-deoxy-ara-uridine complex | | Descriptor: | 1-[3-(4-CARBOXYPIPERIDIN-1-YL)-3-DEOXY-BETA-D-ARABINOFURANOSYL]PYRIMIDINE-2,4(1H,3H)-DIONE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2006-03-03 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The binding of 3'-N-piperidine-4-carboxyl-3'-deoxy-ara-uridine to ribonuclease A in the crystal.
Bioorg.Med.Chem., 14, 2006
|
|
4OE7
 
 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | Descriptor: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
