5LKH
 
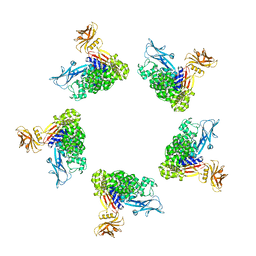 | | Cryo-EM structure of the Tc toxin TcdA1 in its pore state (obtained by flexible fitting) | | Descriptor: | TcdA1 | | Authors: | Gatsogiannis, C, Merino, F, Prumbaum, D, Roderer, D, Leidreiter, F, Meusch, D, Raunser, S. | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Membrane insertion of a Tc toxin in near-atomic detail.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3FB3
 
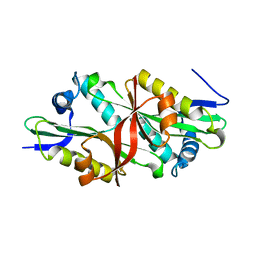 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
6QLF
 
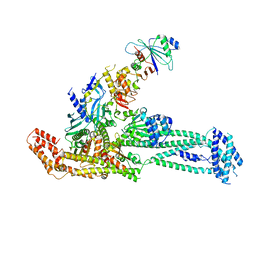 | | Structure of inner kinetochore CCAN complex with mask1 | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
9DIK
 
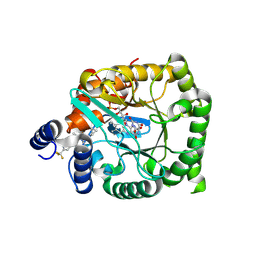 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM681 (N-cyclopropyl-1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxamide) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DIZ
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM959 (6-cyclopropyl-2,4-dimethyl-3-((6-(trifluoromethyl)pyridin-3-yl)methyl)-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, 6-cyclopropyl-2,4-dimethyl-3-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-06 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
8UR5
 
 | |
8UR7
 
 | |
4RVY
 
 | | Serial Time resolved crystallography of Photosystem II using a femtosecond X-ray laser. The S state after two flashes (S3) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.-H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-11-29 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
9FYP
 
 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
9ERX
 
 | | Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor | | Descriptor: | (6aR,10aR)-9-(hydroxymethyl)-6,6-dimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, Antibody ScFv16 Fab fragment, Cannabinoid receptor 1, ... | | Authors: | Thorsen, T.S, Kulkarni, Y, Boggild, A, Drace, T, Nissen, P, Gajhede, M, Boesen, T, Kastrup, J.S, Gloriam, D. | | Deposit date: | 2024-03-25 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of Delta 9 -THC analog activity at the Cannabinoid 1 receptor.
Res Sq, 2024
|
|
5V8G
 
 | | Pekin duck lysozyme isoform I (DEL-I) | | Descriptor: | CHLORIDE ION, THIOCYANATE ION, lysozyme isoform I | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5V3B
 
 | |
1YZ1
 
 | |
8WKY
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
1BMA
 
 | | BENZYL METHYL AMINIMIDE INHIBITOR COMPLEXED TO PORCINE PANCREATIC ELASTASE | | Descriptor: | (1R)-1-benzyl-1-methyl-1-(2-{[4-(1-methylethyl)phenyl]amino}-2-oxoethyl)-2-{(2S)-4-methyl-2-[(trifluoroacetyl)amino]pentanoyl}diazanium, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Peisach, E, Casebier, D, Gallion, S.L, Furth, P, Petsko, G.A, Hogan Jr, J.C, Ringe, D. | | Deposit date: | 1995-05-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of a peptidomimetic aminimide inhibitor with elastase.
Science, 269, 1995
|
|
2Y1O
 
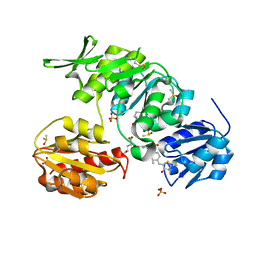 | | Dual-target Inhibitor of MurD and MurE Ligases: Design, Synthesis and Binding Mode Studies | | Descriptor: | (2R)-2-[[3-[[4-[(Z)-(4-OXO-2-SULFANYLIDENE-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]PHENYL]METHYLAMINO]PHENYL]CARBONYLAMINO]PENTANEDIOIC ACID, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Tomasic, T, Sink, R, Kovac, A, Turk, S, Contreras-Martel, C, Dessen, A, Blanot, D, Gobec, S, Zega, A, Kikelj, D, Peterlin-Masic, L. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Dual Inhibitor of MurD and MurE Ligases from Escherichia coli and Staphylococcus aureus.
ACS Med Chem Lett, 3, 2012
|
|
5WYH
 
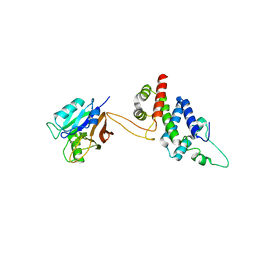 | | Crystal structure of RidL(1-200) complexed with VPS29 | | Descriptor: | GLYCEROL, Interaptin, Vacuolar protein sorting-associated protein 29 | | Authors: | Yao, J, Sun, Q, Jia, D. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanism of inhibition of retromer transport by the bacterial effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BMB
 
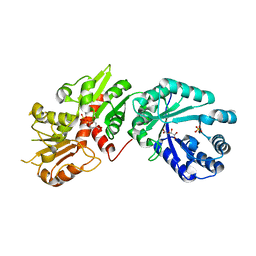 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
6BMQ
 
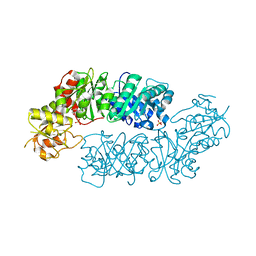 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-09-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
2TRA
 
 | | RESTRAINED REFINEMENT OF TWO CRYSTALLINE FORMS OF YEAST ASPARTIC ACID AND PHENYLALANINE TRANSFER RNA CRYSTALS | | Descriptor: | MAGNESIUM ION, SPERMINE, TRNAASP | | Authors: | Westhof, E, Dumas, P, Moras, D. | | Deposit date: | 1987-11-06 | | Release date: | 1987-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
4WSA
 
 | | Crystal structure of Influenza B polymerase bound to the vRNA promoter (FluB1 form) | | Descriptor: | Influenza B vRNA promoter 3' end, Influenza B vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
6WXO
 
 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 2-histidine 2-glutamate centre TFD-HE | | Descriptor: | GLYCEROL, SULFATE ION, TFD-HE | | Authors: | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6FCM
 
 | | Crystal structure of human PCNA | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Housset, D, Frachet, P. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cytosolic PCNA interacts with p47phox and controls NADPH oxidase NOX2 activation in neutrophils.
J.Exp.Med., 216, 2019
|
|
6X4Q
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 33) | | Descriptor: | (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6X4M
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate (compound 3) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
