7VKH
 
 | |
7VKG
 
 | |
6JHP
 
 | |
8WH5
 
 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHA
 
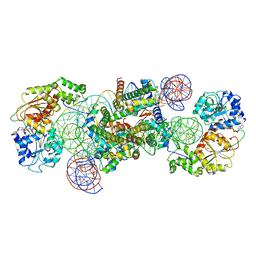 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
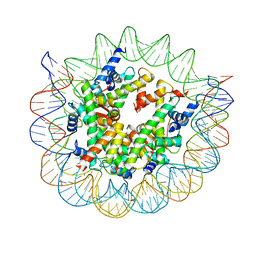 | | Structure of nucleosome core particle of Arabidopsis thaliana | | Descriptor: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH8
 
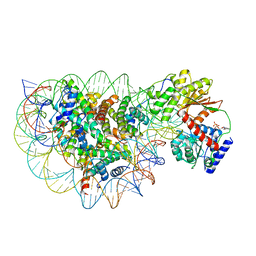 | | Structure of DDM1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
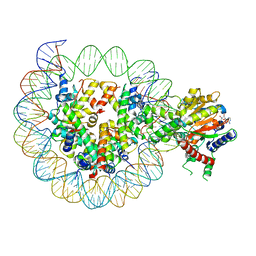 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
7XM0
 
 | | Crystal structure of Sau3AI-C and DNA substrate complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*CP*AP*TP*G)-3'), MAGNESIUM ION, Type II restriction enzyme Sau3AI | | Authors: | Liu, Y, Yu, F, He, J. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of Sau3AI with and without bound DNA suggest a self-activation-based DNA cleavage mechanism.
Structure, 31, 2023
|
|
7XV9
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain | | Descriptor: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV8
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain Homodimer Bound to DR1 Response Element | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), Nuclear receptor subfamily 2 group C member 2, ... | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV6
 
 | |
7XVA
 
 | |
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
5ZEV
 
 | |
7VYW
 
 | |
7VZ2
 
 | | Crystal structure of chromodomain of Arabidopsis LHP1 | | Descriptor: | Chromo domain-containing protein LHP1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Min, J. | | Deposit date: | 2021-11-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the recognition of methylated histone H3 by the Arabidopsis LHP1 chromodomain.
J.Biol.Chem., 298, 2022
|
|
7WDL
 
 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
8H43
 
 | |
8HNI
 
 | | hnRNP A2/B1 RRMs in complex with telomeric DNA | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Liu, Y, Abula, A, Xiao, H, Guo, H, Li, T, Zheng, L, Chen, B, Nguyen, H, Ji, X. | | Deposit date: | 2022-12-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2023
|
|
8IJY
 
 | | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, Deoxyribodipyrimidine photolyase-related protein, ... | | Authors: | Liu, Y, Xu, L, Zhang, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synechococcus elongatus 6-4 photolyase with an 8-HDF as the antenna chromophore and a covalently linked FAD as the catalytic cofactor
To Be Published
|
|
7WDM
 
 | | Fungal immunomodulatory protein FIP-gmi | | Descriptor: | Immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
7DL5
 
 | | Crystal structure of Thermotoga Maritima ferritin mutant at 2.3 Angstrom resolution | | Descriptor: | FE (III) ION, Ferritin, MAGNESIUM ION | | Authors: | Liu, Y, Leng, X, Zang, J, Zhao, G. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A short helix regulates conversion of dimeric and 24-meric ferritin architectures.
Int.J.Biol.Macromol., 203, 2022
|
|
