6R5F
 
 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
6G54
 
 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6GES
 
 | | Crystal structure of ERK1 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 3, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
8QP6
 
 | |
8QP7
 
 | |
1R6H
 
 | | Solution Structure of human PRL-3 | | Descriptor: | protein tyrosine phosphatase type IVA, member 3 isoform 1 | | Authors: | Kozlov, G, Gehring, K, Ekiel, I. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Molecular Function of the Metastasis-associated Phosphatase PRL-3.
J.Biol.Chem., 279, 2004
|
|
4CRL
 
 | |
4F38
 
 | | Crystal structure of geranylgeranylated RhoA in complex with RhoGDI in its active GPPNHP-bound form | | Descriptor: | GERAN-8-YL GERAN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Guo, Z, Tnimov, Z, Alexandrov, K. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Quantitative Analysis of Prenylated RhoA Interaction with Its Chaperone, RhoGDI.
J.Biol.Chem., 287, 2012
|
|
1D5G
 
 | |
5NPH
 
 | |
3MNO
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, F608S) mutations at 1.55A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
4NEU
 
 | | X-ray structure of Receptor Interacting Protein 1 (RIP1)kinase domain with a 1-aminoisoquinoline inhibitor | | Descriptor: | 1-[4-(1-aminoisoquinolin-5-yl)phenyl]-3-(5-tert-butyl-1,2-oxazol-3-yl)urea, MAGNESIUM ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Nolte, R.T, Ward, P, kahler, K.M, Campobasso, N. | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis.
ACS Med Chem Lett, 4, 2013
|
|
5NPI
 
 | |
5NPJ
 
 | |
3MNP
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, V708A, E711G) mutations at 1.50A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3MNE
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by F608S mutation at 1.96A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3O17
 
 | | Crystal Structure of JNK1-alpha1 isoform | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, JIP1, 10MER PEPTIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2010-07-20 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of JNK1-alpha1 isoform
TO BE PUBLISHED
|
|
5HX6
 
 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
1OMD
 
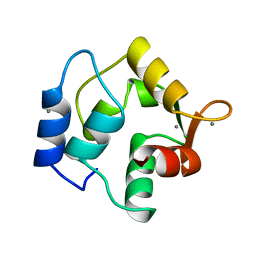 | | STRUCTURE OF ONCOMODULIN REFINED AT 1.85 ANGSTROMS RESOLUTION. AN EXAMPLE OF EXTENSIVE MOLECULAR AGGREGATION VIA CA2+ | | Descriptor: | CALCIUM ION, ONCOMODULIN | | Authors: | Ahmed, F.R, Przybylska, M, Rose, D.R, Birnbaum, G.I, Pippy, M.E, Macmanus, J.P. | | Deposit date: | 1990-04-19 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oncomodulin refined at 1.85 A resolution. An example of extensive molecular aggregation via Ca2+.
J.Mol.Biol., 216, 1990
|
|
3O2M
 
 | |
1DCC
 
 | |
6DMI
 
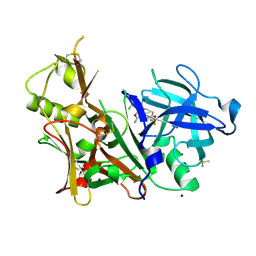 | | A multiconformer ligand model of 5T5 bound to BACE-1 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
3PAZ
 
 | | REDUCED NATIVE PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-20 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
5I2R
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C2(=NN(c1cccc(c1)OC(F)(F)F)C=CC2=O)c3ccnn3c4ccccc4, micromolar IC50=0.019462 | | Descriptor: | 3-(1-phenyl-1H-pyrazol-5-yl)-1-[3-(trifluoromethoxy)phenyl]pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Rudolph, M.G. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
