2W2Q
 
 | | PCSK9-deltaC D374H mutant bound to WT EGF-A of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
6ESM
 
 | | Crystal structure of MMP9 in complex with inhibitor BE4. | | Descriptor: | (2~{S})-2-[2-[4-(4-methoxyphenyl)phenyl]sulfanylphenyl]pentanedioic acid, CALCIUM ION, Matrix metalloproteinase-9,Matrix metalloproteinase-9, ... | | Authors: | Ciccone, L, Tepshi, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-10-23 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
379D
 
 | | THE STRUCTURAL BASIS OF HAMMERHEAD RIBOZYME SELF-CLEAVAGE | | Descriptor: | COBALT (II) ION, RNA (5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*CP*UP*CP*GP*UP*AP*AP*GP*A P*GP*UP*CP*AP*CP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3') | | Authors: | Murray, J.B, Terwey, D.P, Maloney, L, Karpeisky, A, Usman, N, Beigelman, L, Scott, W.G. | | Deposit date: | 1998-02-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of hammerhead ribozyme self-cleavage.
Cell(Cambridge,Mass.), 92, 1998
|
|
2V09
 
 | | SENS161-164DSSN mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Burrell, M.R, Just, V.J, Bowater, L, Fairhurst, S.A, Requena, L, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxalate Decarboxylase and Oxalate Oxidase Activities Can be Interchanged with a Specificity Switch of Up to 282 000 by Mutating an Active Site Lid.
Biochemistry, 46, 2007
|
|
2VM5
 
 | | HUMAN BIR2 DOMAIN OF BACULOVIRAL INHIBITOR OF APOPTOSIS REPEAT- CONTAINING 1 (BIRC1) | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 1, GLYCEROL, ZINC ION | | Authors: | Herman, M.D, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-23 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Bir Domains from Human Naip and Ciap2.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1FD8
 
 | | SOLUTION STRUCTURE OF THE CU(I) FORM OF THE YEAST METALLOCHAPERONE, ATX1 | | Descriptor: | ATX1 COPPER CHAPERONE, COPPER (I) ION | | Authors: | Arnesano, F, Banci, L, Bertini, I, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-07-20 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1.
Biochemistry, 40, 2001
|
|
2V4U
 
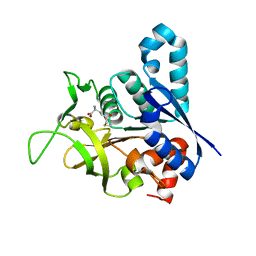 | | Human CTP synthetase 2 - glutaminase domain in complex with 5-OXO-L- NORLEUCINE | | Descriptor: | CTP SYNTHASE 2 | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wisniewska, M, Weigelt, J, Wikstrom, M, Nordlund, P. | | Deposit date: | 2008-09-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human Ctp Synthetase 2 - Glutaminase Domain in Complex with 5-Oxo-L-Norleucine
To be Published
|
|
8GCM
 
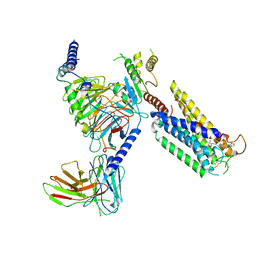 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (5S)-5-[(3R)-4,4-difluoro-3-hydroxy-4-phenylbutyl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GCP
 
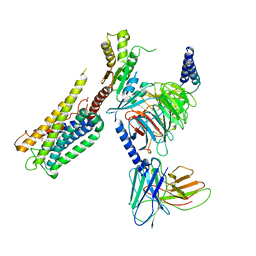 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2W04
 
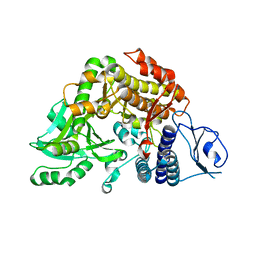 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with citrate in ATP binding site from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, CITRATE ANION | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acsd Catalyzes Enantioselective Citrate Desymmetrization in Siderophore Biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
2MYM
 
 | |
1FES
 
 | | SOLUTION STRUCTURE OF THE APO FORM OF THE YEAST METALLOCHAPERONE, ATX1 | | Descriptor: | ATX1 COPPER CHAPERONE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-07-22 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1.
Biochemistry, 40, 2001
|
|
2Y71
 
 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-1,4,5-trihydroxy-3-((5-methylbenzo(b) thiophen-2-yl)methoxy)cyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-1,4,5-trihydroxy-3-[(5-methyl-1-benzothiophen-2-yl)methoxy]cyclohex-2-ene-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
8DYP
 
 | |
2MYL
 
 | |
2Y76
 
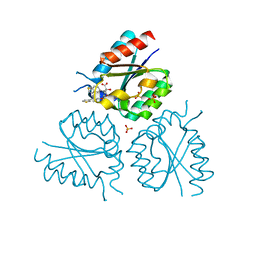 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-5-ylmethoxy)-2-(benzo(b) thiophen-5-ylmethyl)-1,4,5-trihydroxycyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[b]THIOPHEN-5-YL)METHOXY-2-(BENZO[b]THIOPHEN-5-YL)METHYL-1,4,5-TRIHYDROXYCYCLOHEX-2-ENE-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
6N45
 
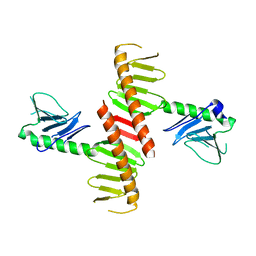 | |
2Y77
 
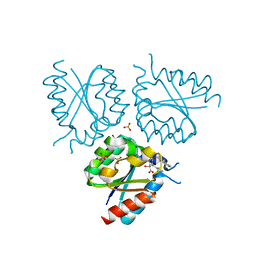 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-2-ylmethoxy)-1,4,5- trihydroxy-2-(thiophen-2-ylmethyl)cyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[B]THIOPHEN-2-YL)METHOXY-1,4,5-TRIHYDROXY-2-(THIEN-2-YL)METHYLCYCLOHEX-2-EN-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
8DW9
 
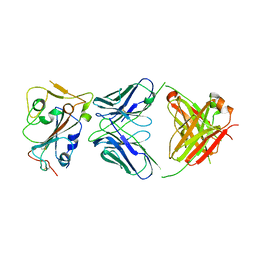 | |
8DXS
 
 | |
1GK5
 
 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
5YMS
 
 | |
8DWA
 
 | |
3BID
 
 | | Crystal structure of the NMB1088 protein from Neisseria meningitidis. Northeast Structural Genomics Consortium target MR91 | | Descriptor: | UPF0339 protein NMB1088 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, L, Fang, Y, Xiao, R, Owen, L.A, Maglaqui, M, Cunningham, K, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NMB1088 protein from Neisseria meningitidis.
To be Published
|
|
3BB6
 
 | | Crystal structure of the P64488 protein from E.coli (strain K12). Northeast Structural Genomics Consortium target ER596 | | Descriptor: | Uncharacterized protein yeaR, ZINC ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray structure of the P64488 from E.coli (strain K12).
To be Published
|
|
