8J9Z
 
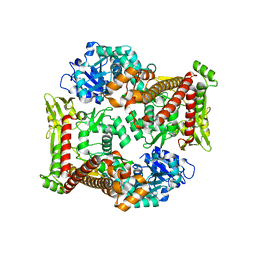 | |
8J9Y
 
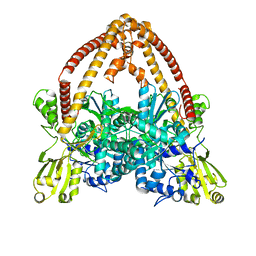 | |
5XS9
 
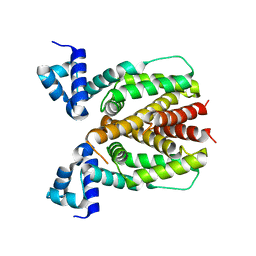 | | Crystal structure of Mycobacterium smegmatis BioQ | | Descriptor: | TetR family transcriptional regulator | | Authors: | Zhang, Y, Ji, Q, Feng, Y. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of BioQ suggests a distinct regulatory mechanism for biotin, a nutritional virulence factor in Mycobacterium
To Be Published
|
|
1FHT
 
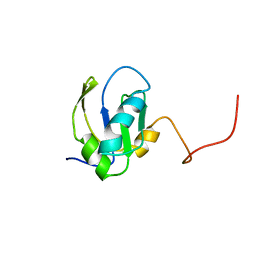 | | RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1996-02-21 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding.
J.Mol.Biol., 257, 1996
|
|
5W8P
 
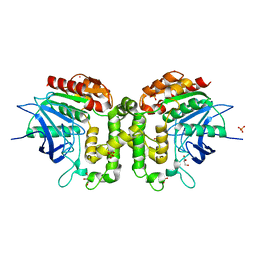 | | Homoserine transacetylase MetX from Mycobacterium abscessus | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase, PHOSPHATE ION, ... | | Authors: | Rodriguez, E.S, Reed, R.W, Korotkov, K.V. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of mycobacterial homoserine transacetylases central to methionine biosynthesis reveals druggable active site.
Sci Rep, 9, 2019
|
|
5Y7R
 
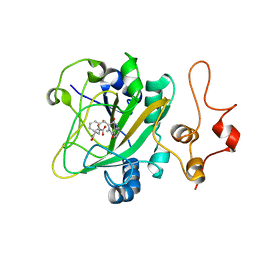 | | Quaternary complex of AsqJ-Fe3+-2OG-cyclopeptin | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, ... | | Authors: | Liao, H.J, Lin, T.S. | | Deposit date: | 2017-08-17 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Insights into the Desaturation of Cyclopeptin and its C3 Epimer Catalyzed by a non-Heme Iron Enzyme: Structural Characterization and Mechanism Elucidation.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5W4W
 
 | |
5YUP
 
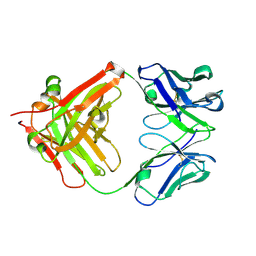 | | Crystal Structure of the Fab fragment of FVIIa antibody mAb4F5 | | Descriptor: | the heavy chain of the Fab fragment of FVIIa antibody mAb4F5, the light chain of the Fab fragment of FVIIa antibody mAb4F5 | | Authors: | Jiang, L.G, Persson, E, Huang, M.D. | | Deposit date: | 2017-11-22 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure, epitope, and functional impact of an antibody against a superactive FVIIa provide insights into allosteric mechanism.
Res Pract Thromb Haemost, 3, 2019
|
|
5Y7T
 
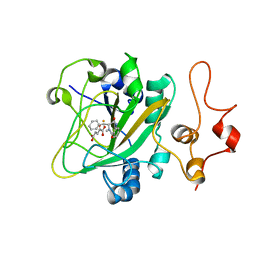 | | Quaternary complex of AsqJ-Fe3+-2OG-D-cyclopeptin | | Descriptor: | (3R)-4-methyl-3-(phenylmethyl)-1,3-dihydro-1,4-benzodiazepine-2,5-dione, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Liao, H.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the Desaturation of Cyclopeptin and its C3 Epimer Catalyzed by a non-Heme Iron Enzyme: Structural Characterization and Mechanism Elucidation.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5ZB2
 
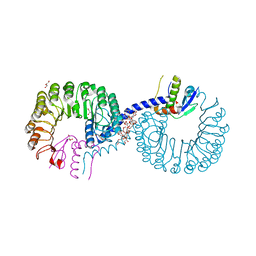 | | Crystal structure of Rad7 and Elc1 complex in yeast | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Jiang, T, Liu, L, Huo, Y. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.30000544 Å) | | Cite: | Crystal structure of the yeast Rad7-Elc1 complex and assembly of the Rad7-Rad16-Elc1-Cul3 complex.
DNA Repair (Amst.), 77, 2019
|
|
7BX8
 
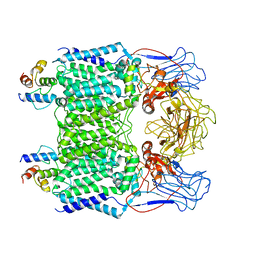 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | Descriptor: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7BWR
 
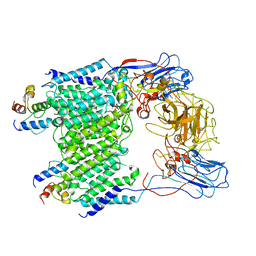 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7CNL
 
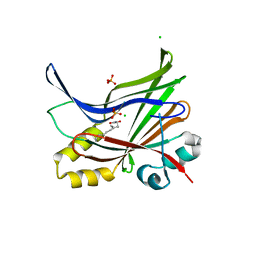 | | Crystal structure of TEAD3 in complex with VT105 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, N-[(1S)-1-pyridin-2-ylethyl]-8-[4-(trifluoromethyl)phenyl]quinoline-3-carboxamide, ... | | Authors: | Tang, T.T, Konradi, A.W. | | Deposit date: | 2020-08-01 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small Molecule Inhibitors of TEAD Auto-palmitoylation Selectively Inhibit Proliferation and Tumor Growth of NF2 -deficient Mesothelioma.
Mol.Cancer Ther., 20, 2021
|
|
7CIP
 
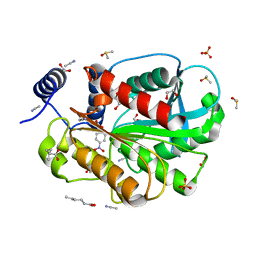 | | Microbial Hormone-sensitive lipase E53 wild type | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 wild type
To Be Published
|
|
7D9P
 
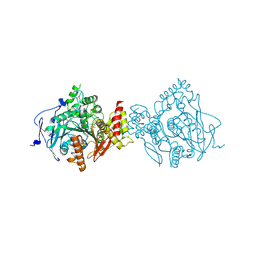 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 12 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(2-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9O
 
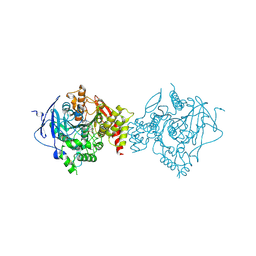 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 2 | | Descriptor: | (2R)-2-[[4-fluoranyl-1-[(4-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9Q
 
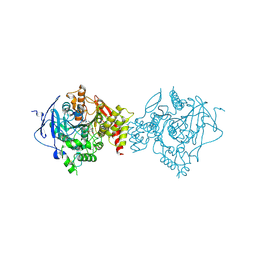 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 7 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(3-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D2F
 
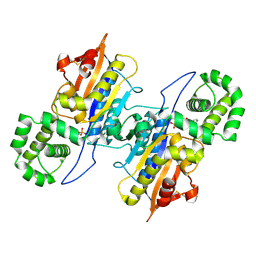 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Major acid phosphatase | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7D57
 
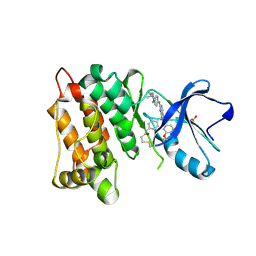 | | C-Src in complex with FIIN-2 | | Descriptor: | N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7D5O
 
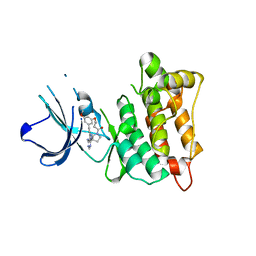 | | C-Src in complex with TAS-120 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2020-09-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7DOQ
 
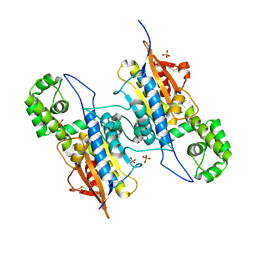 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | Acid phosphatase, PHOSPHATE ION | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-12-16 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7BIP
 
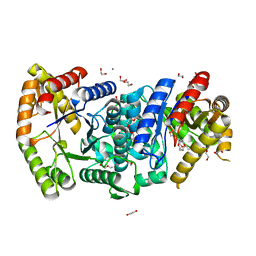 | | Crystal structure of monooxygenase RslO1 from Streptomyces bottropensis | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zhang, L, Zuo, C, Bechthold, A, Einsle, O. | | Deposit date: | 2021-01-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biosynthesis of the Tricyclic Aromatic Type II Polyketide Rishirilide: New Potential Third Ring Oxygenation after Three Cyclization Steps.
Mol Biotechnol., 63, 2021
|
|
7BIO
 
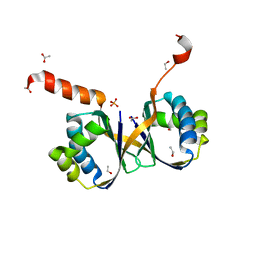 | | Crystal structure of monooxygenase RslO4 from Streptomyces bottropensis | | Descriptor: | ACETATE ION, ETHANOL, Monooxygenase/putative anthronoxygenase, ... | | Authors: | Zhang, L, Zuo, C, Alali, A, Bechthold, A, Einsle, O. | | Deposit date: | 2021-01-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Biosynthesis of the Tricyclic Aromatic Type II Polyketide Rishirilide: New Potential Third Ring Oxygenation after Three Cyclization Steps.
Mol Biotechnol., 63, 2021
|
|
7EN6
 
 | | The crystal structure of Escherichia coli MurR in apo form | | Descriptor: | HTH-type transcriptional regulator MurR, PHOSPHATE ION | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7EN5
 
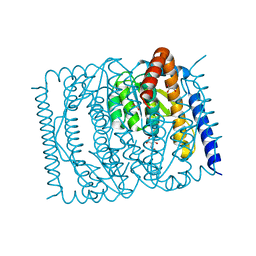 | | The crystal structure of Escherichia coli MurR in complex with N-acetylglucosamine-6-phosphate | | Descriptor: | 2-METHOXYETHANOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhang, Y, Chen, W, Ji, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
