7T9U
 
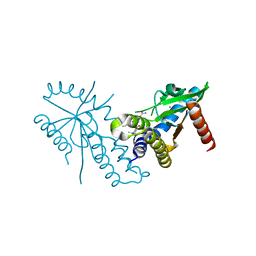 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
2P8S
 
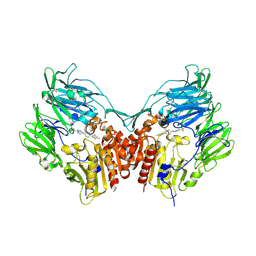 | | Human dipeptidyl peptidase IV/CD26 in complex with a cyclohexalamine inhibitor | | Descriptor: | (1S,2R,5S)-5-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-2-(2,4,5-TRIFLUOROPHENYL)CYCLOHEXANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a novel, potent, and orally bioavailable cyclohexylamine DPP-4 inhibitor by application of molecular modeling and X-ray crystallography of sitagliptin
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3W95
 
 | |
2L42
 
 | |
4QO8
 
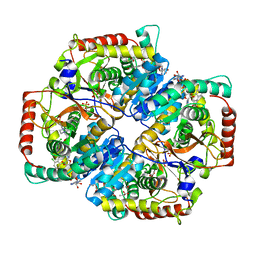 | | Lactate Dehydrogenase A in complex with substituted 3-Hydroxy-2-mercaptocyclohex-2-enone compound 104 | | Descriptor: | (5S)-2-[(2-chlorophenyl)sulfanyl]-5-(2,6-dichlorophenyl)-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Identification of substituted 3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2L7E
 
 | |
3GXB
 
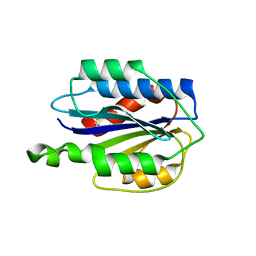 | | Crystal structure of VWF A2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Zhou, Y.F, Springer, T.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4QO7
 
 | |
4R68
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 31 | | Descriptor: | (1S)-1-phenylethyl (4-chloro-3-{[(4S)-4-(2,6-dichlorophenyl)-2-hydroxy-6-oxocyclohex-1-en-1-yl]sulfanyl}phenyl)acetate, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
2L83
 
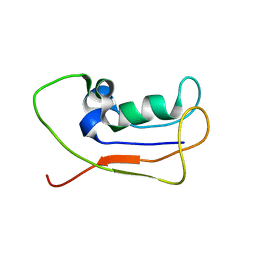 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
7WNT
 
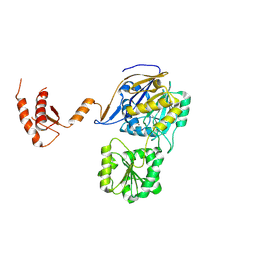 | | RNase J from Mycobacterium tuberculosis | | Descriptor: | Ribonuclease J, ZINC ION | | Authors: | Li, J, Bao, L, Hu, J, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
8TXG
 
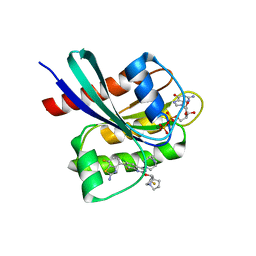 | | Crystal structure of KRAS G12D in complex with GDP and compound 8 | | Descriptor: | (4M)-4-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-7-fluoro-1,3-benzothiazol-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8TXH
 
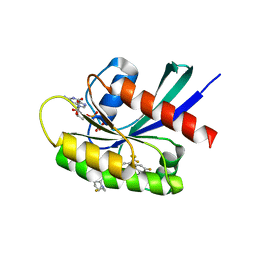 | | Crystal structure of KRAS G12D in complex with GDP and compound 14 | | Descriptor: | (4P)-2-amino-4-{4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}-6-(trifluoromethyl)quinazolin-7-yl}-7-fluoro-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8TXE
 
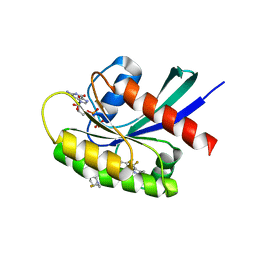 | | Crystal structure of KRAS G12D in complex with GDP and compound 5 | | Descriptor: | (6M)-6-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-4-methyl-5-(trifluoromethyl)pyridin-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
5XGH
 
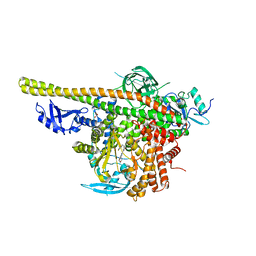 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
4R69
 
 | | Lactate Dehydrogenase in complex with inhibitor compound 13 | | Descriptor: | (5R)-2-[(2-chlorophenyl)sulfanyl]-5-[2,6-dichloro-3-(tetrahydro-2H-pyran-4-ylamino)phenyl]-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Optimization of 5-(2,6-dichlorophenyl)-3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 25, 2014
|
|
5GNF
 
 | | Crystal structure of anti-CRISPR protein AcrF3 | | Descriptor: | CALCIUM ION, Uncharacterized protein AcrF3 | | Authors: | Wang, J, Wang, Y. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
5XJN
 
 | | cytochrome P450 CREJ in complex with (4-ethylphenyl) dihydrogen phosphate | | Descriptor: | (4-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S, Feng, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4RE1
 
 | | Crystal structure of human TEAD1 and disulfide-engineered YAP | | Descriptor: | CHLORIDE ION, Transcriptional enhancer factor TEF-1, Yorkie homolog | | Authors: | Xu, Z, Zhou, Z. | | Deposit date: | 2014-09-21 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting Hippo pathway by specific interruption of YAP-TEAD interaction using cyclic YAP-like peptides.
Faseb J., 29, 2015
|
|
1D6E
 
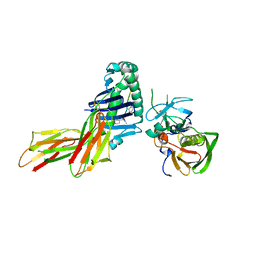 | | CRYSTAL STRUCTURE OF HLA-DR4 COMPLEX WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, PEPTIDOMIMETIC INHIBITOR | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-13 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
6PIT
 
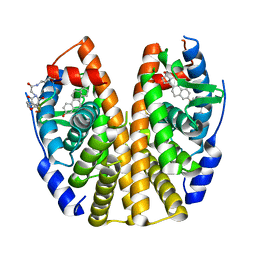 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with SRC2 Stapled Peptide 41A and Estradiol | | Descriptor: | ESTRADIOL, Estrogen receptor, Stapled Peptide 41A | | Authors: | Fanning, S.W, Montgomery, J.E, Greene, G.L, Moellering, R.E. | | Deposit date: | 2019-06-27 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Versatile Peptide Macrocyclization with Diels-Alder Cycloadditions.
J.Am.Chem.Soc., 141, 2019
|
|
7V6Y
 
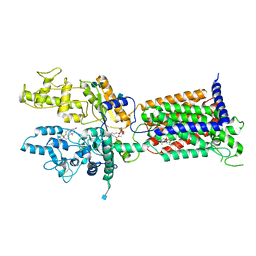 | | Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.5 angstrom (re-processed with dataset of 7dzq) | | Descriptor: | (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
7V6Z
 
 | | Cryo-EM structure of Patched1 (V1084A mutant) in lipid nanodisc, 3.64 angstrom (reprocessed with the dataset of 7dzp) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
