6TS6
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-5-yl]-5-(2-cyanopropan-2-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
5X1E
 
 | |
8IVU
 
 | | Crystal Structure of Human NAMPT in complex with A4276 | | Descriptor: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kang, B.G, Cha, S.S. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09000921 Å) | | Cite: | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
5G54
 
 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2C
 
 | | The crystal structure of light-driven chloride pump ClR (T102D) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2A
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0 with Bromide ion. | | Descriptor: | BROMIDE ION, CHLORIDE PUMPING RHODOPSIN, RETINAL | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
7YC5
 
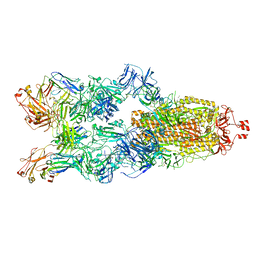 | | Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from K202.B, bispecific antibody, ... | | Authors: | Yoo, Y, Cho, H.S. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Novel bispecific human antibody platform specifically targeting a fully open spike conformation potently neutralizes multiple SARS-CoV-2 variants.
Antiviral Res., 212, 2023
|
|
6TL8
 
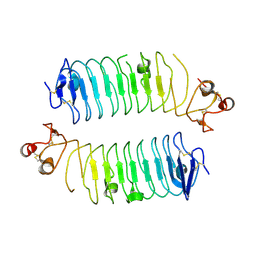 | | Structural basis of SALM3 dimerization and adhesion complex formation with the presynaptic receptor protein tyrosine phosphatases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Myeloid cell surface antigen CD33,Leucine-rich repeat and fibronectin type-III domain-containing protein 4 | | Authors: | Karki, S, Shkumatov, A.V, Bae, S, Ko, J, Kajander, T. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of SALM3 dimerization and synaptic adhesion complex formation with PTP sigma.
Sci Rep, 10, 2020
|
|
5C16
 
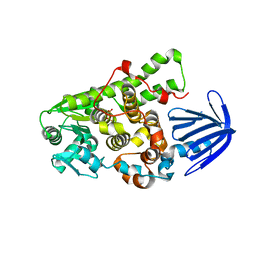 | | Myotubularin-related proetin 1 | | Descriptor: | Myotubularin-related protein 1, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2015-06-13 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Plos One, 11, 2016
|
|
7XW2
 
 | |
7XW3
 
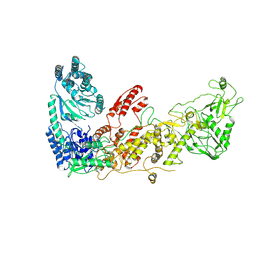 | |
8J26
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-4, AM047-6, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8J1Q
 
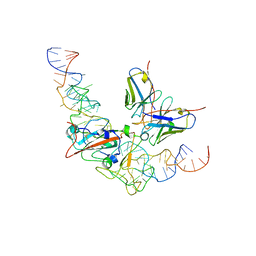 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
6W30
 
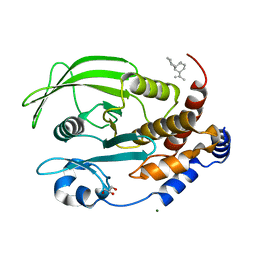 | | Protein Tyrosine Phosphatase 1B Bound to Amorphadiene | | Descriptor: | Amorphadiene, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J. | | Deposit date: | 2020-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
2G97
 
 | |
8X9G
 
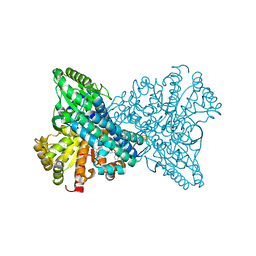 | | Crystal structure of CO dehydrogenase mutant in complex with BV | | Descriptor: | 1-(phenylmethyl)-4-[1-(phenylmethyl)pyridin-1-ium-4-yl]pyridin-1-ium, Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9H
 
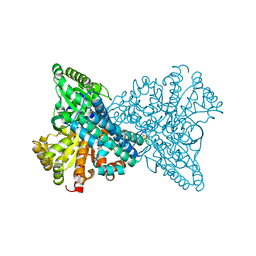 | | Crystal structure of CO dehydrogenase mutant (F41C) | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9F
 
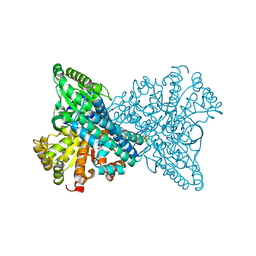 | | Crystal structure of CO dehydrogenase mutant in complex with EV | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-4-(1-ethylpyridin-1-ium-4-yl)pyridin-1-ium, Carbon monoxide dehydrogenase 2, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9E
 
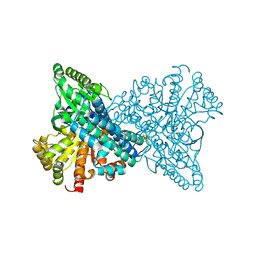 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in low PEG concentration | | Descriptor: | 1,2-ETHANEDIOL, Carbon monoxide dehydrogenase 2, FE (III) ION, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9D
 
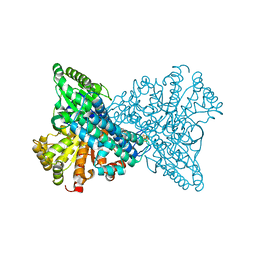 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in high PEG concentration | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
2G96
 
 | |
2G95
 
 | |
6IWD
 
 | | The PTP domain of human PTPN14 in a complex with the CR3 domain of HPV18 E7 | | Descriptor: | CHLORIDE ION, HPV18 E7, PHOSPHATE ION, ... | | Authors: | Yun, H.-Y, Kim, S.J, Ku, B. | | Deposit date: | 2018-12-05 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus.
Plos Biol., 17, 2019
|
|
