3EWS
 
 | | Human DEAD-box RNA-helicase DDX19 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B | | Authors: | Lehtio, l, Karlberg, t, Andersson, j, Arrowsmith, c.h, Berglund, h, Bountra, c, Collins, r, Dahlgren, l.g, Edwards, a.m, Flodin, s, Flores, a, Graslund, s, Hammarstrom, m, Johansson, a, Johansson, i, Kotenyova, t, Moche, m, Nilsson, m.e, Nordlund, p, Nyman, t, Olesen, k, Persson, c, Sagemark, j, Thorsell, a.g, Tresaugues, l, Van den berg, s, Weigelt, j, Welin, m, Wikstrom, m, Wisniewska, m, Schueler, h, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The DEXD/H-box RNA Helicase DDX19 Is Regulated by an {alpha}-Helical Switch.
J.Biol.Chem., 284, 2009
|
|
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
7K6H
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to XMD8-92 | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Karim, M.R, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2020-09-20 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7K6G
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to ERK5-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-2-({2-ethoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Karim, M.R, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2020-09-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
6J9J
 
 | |
7W5U
 
 | | Acetyl-CoA Carboxylase-AccB | | Descriptor: | Acetyl-CoA carboxylase complex, beta-chain, GLYCEROL, ... | | Authors: | Ali, I, Zheng, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of Acetyl-CoA carboxylase (AccB) from Streptomyces antibioticus and insights into the substrate-binding through in silico mutagenesis and biophysical investigations.
Comput Biol Med, 145, 2022
|
|
6B9K
 
 | |
7KO0
 
 | | Crystal structure of the second bromodomain (BD2) of human BRD4 bound to SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Karim, M.R, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2020-11-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
5TX2
 
 | | Miniature TGF-beta2 3-mutant monomer | | Descriptor: | Transforming growth factor beta-2 | | Authors: | Taylor, A.B, Kim, S.K, Hart, P.J, Hinck, A.P. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
5Z35
 
 | | Structure of Y68F mutant metal free periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, Periplasmic solute binding protein, ... | | Authors: | Saini, G, Sharma, N, Dalal, V, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-01-05 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The analysis of subtle internal communications through mutation studies in periplasmic metal uptake protein CLas-ZnuA2
J. Struct. Biol., 204, 2018
|
|
3FO5
 
 | | Human START domain of Acyl-coenzyme A thioesterase 11 (ACOT11) | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van-Den-Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Shueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
7L72
 
 | | Crystal structure of the second bromodomain (BD2) of human BRD3 bound to Ro3280 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M, Schonbrunn, E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
3FE2
 
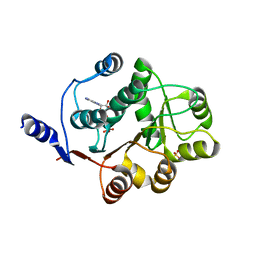 | | Human DEAD-BOX RNA helicase DDX5 (P68), conserved domain I in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX5, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases
Plos One, 5, 2010
|
|
3G0H
 
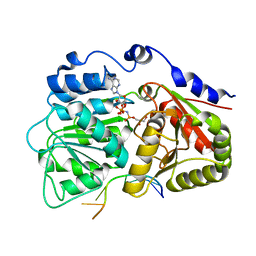 | | Human dead-box RNA helicase DDX19, in complex with an ATP-analogue and RNA | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase DDX19B, GLYCEROL, ... | | Authors: | Karlberg, T, Lehtio, L, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The DEXD/H-box RNA Helicase DDX19 Is Regulated by an {alpha}-Helical Switch.
J.Biol.Chem., 284, 2009
|
|
5Z2J
 
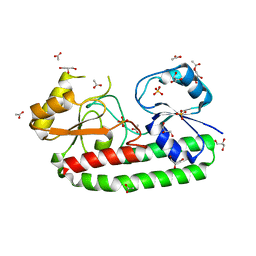 | | structure of S38A mutant metal-free periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, Periplasmic solute binding protein, ... | | Authors: | Saini, G, Sharma, N, Dalal, V, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The analysis of subtle internal communications through mutation studies in periplasmic metal uptake protein CLas-ZnuA2
J. Struct. Biol., 204, 2018
|
|
5ZHA
 
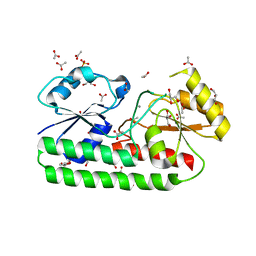 | | Structure of Y68F mutant Mn-Bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Saini, G, Sharma, N, Dalal, V, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-03-12 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The analysis of subtle internal communications through mutation studies in periplasmic metal uptake protein CLas-ZnuA2
J. Struct. Biol., 204, 2018
|
|
5N2B
 
 | |
5VAE
 
 | |
3BXM
 
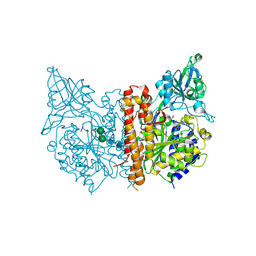 | | Structure of an inactive mutant of human glutamate carboxypeptidase II [GCPII(E424A)] in complex with N-acetyl-Asp-Glu (NAAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Barinka, C. | | Deposit date: | 2008-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reaction mechanism of glutamate carboxypeptidase II revealed by mutagenesis, X-ray crystallography, and computational methods.
Biochemistry, 48, 2009
|
|
5VAF
 
 | |
7KP6
 
 | | Structure of Ack1 kinase in complex with a selective inhibitor | | Descriptor: | 5-chloro-N~2~-[4-(4-methylpiperazin-1-yl)phenyl]-N~4~-{[(2R)-oxolan-2-yl]methyl}pyrimidine-2,4-diamine, Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Thakur, M.K, Miller, W.T, Mahajan, N, Seeliger, M.A. | | Deposit date: | 2020-11-10 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance.
Nat Commun, 13, 2022
|
|
3DKP
 
 | | Human DEAD-box RNA-helicase DDX52, conserved domain I in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lehtio, L, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Wisniewska, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3UMQ
 
 | | Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, Peptidoglycan recognition protein 1, butanoic acid | | Authors: | Pandey, N, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
5Z2K
 
 | | Structure of S38A mutant Mn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Saini, G, Sharma, N, Dalal, V, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The analysis of subtle internal communications through mutation studies in periplasmic metal uptake protein CLas-ZnuA2
J. Struct. Biol., 204, 2018
|
|
4S10
 
 | |
