3C3J
 
 | | Crystal structure of tagatose-6-phosphate ketose/aldose isomerase from Escherichia coli | | Descriptor: | Putative tagatose-6-phosphate ketose/aldose isomerase | | Authors: | Zhang, R, Skarina, T, Egorova, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the tagatose-6-phosphate ketose/aldose isomerase from Escherichia coli.
To be Published
|
|
3BNI
 
 | | Crystal structure of TetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | Putative TetR-family transcriptional regulator, TETRAETHYLENE GLYCOL | | Authors: | Osipiuk, J, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of TetR-family transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
3BQY
 
 | | Crystal structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2). | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Putative TetR family transcriptional regulator | | Authors: | Cuff, M.E, Skarina, T, Kagan, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a possible TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
TO BE PUBLISHED
|
|
3C07
 
 | | Crystal structure of a TetR family transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | Putative tetR-family transcriptional regulator, SULFATE ION | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a TetR family transcriptional regulator from Streptomyces coelicolor A3(2).
To be Published
|
|
3BRQ
 
 | | Crystal structure of the Escherichia coli transcriptional repressor ascG | | Descriptor: | HTH-type transcriptional regulator ascG, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the E. coli transcriptional repressor ascG.
To be Published
|
|
3C2B
 
 | | Crystal structure of TetR transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | FORMIC ACID, Transcriptional regulator, TetR family | | Authors: | Osipiuk, J, Skarina, T, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TetR transcriptional regulator from Agrobacterium tumefaciens.
To be Published
|
|
1R7L
 
 | | 2.0 A Crystal Structure of a Phage Protein from Bacillus cereus ATCC 14579 | | Descriptor: | Phage protein | | Authors: | Zhang, R, Joachimiak, G, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of phage protein BC1872 from Bacillus cereus, a singleton with new fold
Proteins, 64, 2006
|
|
6NIO
 
 | | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, G, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis
To Be Published
|
|
8VZK
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase CfhHACS from Chloroflexi bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Gade, P, Mayfield, A, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
5BMZ
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA. | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*AP*TP*CP*AP*GP*TP*TP*AP*AP*AP*CP*TP*GP*AP*TP*AP*TP*TP*C)-3'), HcaR protein | | Authors: | Kim, Y, Joachimiak, G, Biglow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA.
To Be Published
|
|
5TF0
 
 | | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 3 N-terminal domain protein, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.2021 Å) | | Cite: | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis
To Be Published, 2016
|
|
5T86
 
 | | Crystal structure of CDI complex from E. coli A0 34/86 | | Descriptor: | ACETATE ION, CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
6BB9
 
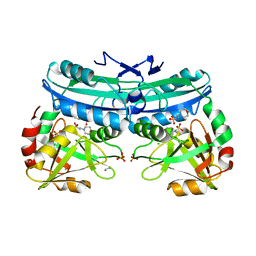 | | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-4-deoxychorismate lyase, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-17 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2
To Be Published
|
|
6AWA
 
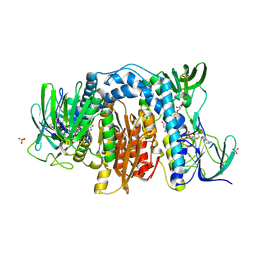 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6AZI
 
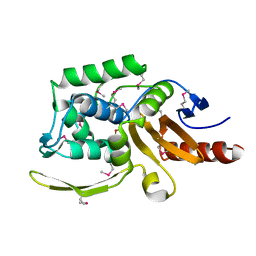 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | Authors: | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-09-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
6WLC
 
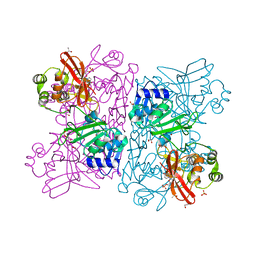 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
7S6O
 
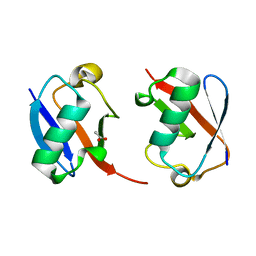 | | The crystal structure of Lys48-linked di-ubiquitin | | Descriptor: | ACETATE ION, Ubiquitin | | Authors: | Osipiuk, J, Tesar, C, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
6B6L
 
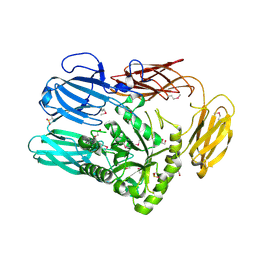 | | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Glycosyl hydrolase family 2, ... | | Authors: | Tan, K, Joachimiak, G, Nocek, B, Enddres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
6U8J
 
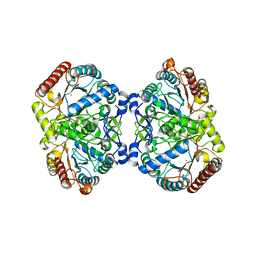 | | Crystal structure of 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase/phospho-2-dehydro-3-deoxyheptonate aldolase (Aro3) from Candida auris | | Descriptor: | Phospho-2-dehydro-3-deoxyheptonate aldolase, UNKNOWN ATOM OR ION | | Authors: | Michalska, K, Evdokimova, E, Semper, C, Di Leo, R, Stogios, P.J, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Crystal structure of 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase/phospho-2-dehydro-3-deoxyheptonate aldolase (Aro3) from
Candida auris
To Be Published
|
|
8EP6
 
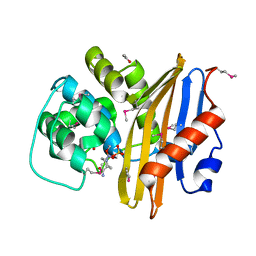 | | Crystal Structure of the Beta-lactamase Class D from Chitinophaga pinensis in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, ACETIC ACID, Beta-lactamase Class D Cpin_0907 | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Beta-lactamase Class D from Chitinophaga pinensis in the complex with Avibactam.
To Be Published
|
|
8EP7
 
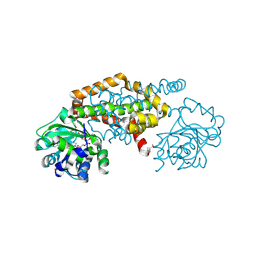 | | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in complex with NADP | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase (NADP(+)) 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Maltseva, N, Osipiuk, J, Gu, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in the complex with NADP.
To Be Published
|
|
1WRP
 
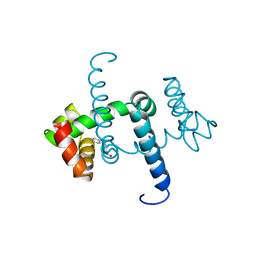 | | FLEXIBILITY OF THE DNA-BINDING DOMAINS OF TRP REPRESSOR | | Descriptor: | TRP REPRESSOR, TRYPTOPHAN | | Authors: | Schewitz, R.W, Otwinowski, Z, Lawson, C.L, Joachimiak, A, Sigler, P.B. | | Deposit date: | 1987-12-01 | | Release date: | 1988-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility of the DNA-binding domains of trp repressor.
Proteins, 3, 1988
|
|
7S6P
 
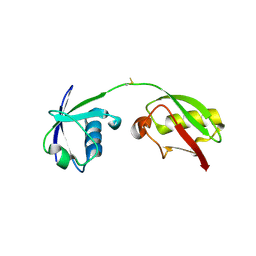 | | The crystal structure of human ISG15 | | Descriptor: | Ubiquitin-like protein ISG15 | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
6U83
 
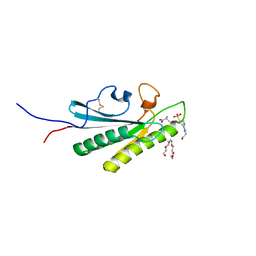 | | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-ALANINE, Outer membrane associated protein, ... | | Authors: | Michalska, K, Skarina, T, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3566 Å) | | Cite: | OmpA-like domain of FopA1 from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
