3J2M
 
 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the extended T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
7KES
 
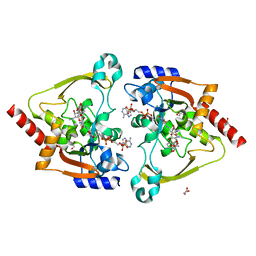 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in complex with apramycin and CoA | | Descriptor: | APRAMYCIN, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-12 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
1WPB
 
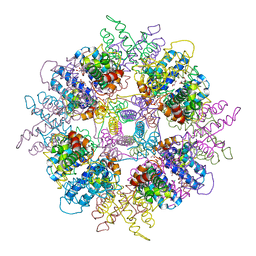 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
3J47
 
 | | Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid | | Descriptor: | 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN12, 26S proteasome regulatory subunit RPN3, ... | | Authors: | Estrin, E, Lopez-Blanco, J.R, Chacon, P, Martin, A. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Formation of an Intricate Helical Bundle Dictates the Assembly of the 26S Proteasome Lid.
Structure, 21, 2013
|
|
1UH1
 
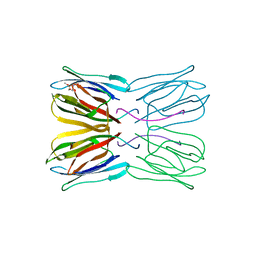 | | Crystal structure of jacalin- GalNAc-beta(1-3)-Gal-alpha-O-Me complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-methyl alpha-D-galactopyranoside, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
8EYP
 
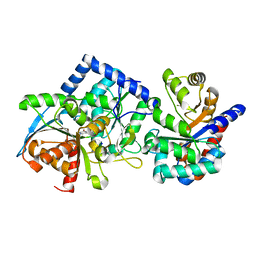 | | Joint X-ray/neutron structure of Salmonella typhimurium tryptophan synthase internal aldimine from microgravity-grown crystal | | Descriptor: | SODIUM ION, Tryptophan synthase alpha chain, Tryptophan synthase beta chain | | Authors: | Drago, V.N, Kovalevsky, A, Blakeley, M.P, Forsyth, V.T, Mueser, T.C. | | Deposit date: | 2022-10-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Neutron diffraction from a microgravity-grown crystal reveals the active site hydrogens of the internal aldimine form of tryptophan synthase.
Cell Rep Phys Sci, 5, 2024
|
|
8EJV
 
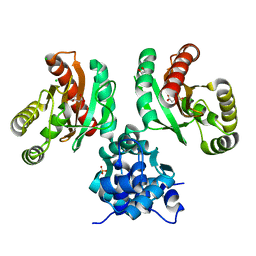 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR in complex with succinate
To Be Published
|
|
3SJG
 
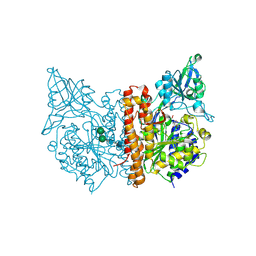 | | Human glutamate carboxypeptidase II (E424A inactive mutant ) in complex with N-acetyl-aspartyl-aminooctanoic acid | | Descriptor: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
8EJU
 
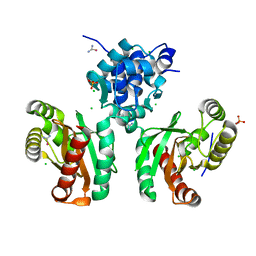 | | The crystal structure of Pseudomonas putida PcaR | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transcription regulatory protein (Pca regulon), ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR
To Be Published
|
|
2NNN
 
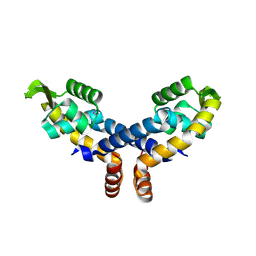 | | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Altamentova, S, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of probable transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
3SJE
 
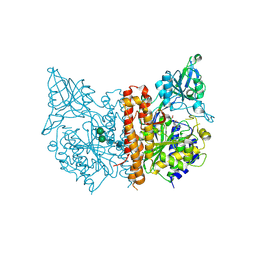 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-aminononanoic acid | | Descriptor: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
1WWE
 
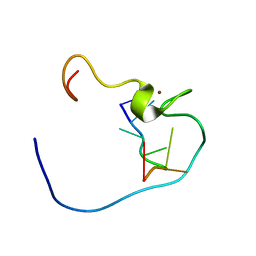 | | NMR Structure Determined for MLV NC complex with RNA Sequence UUUUGCU | | Descriptor: | 5'-R(P*UP*UP*UP*UP*GP*CP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
2NPT
 
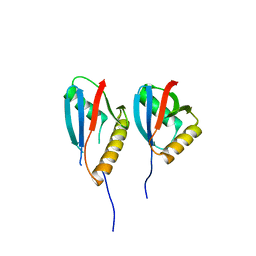 | | Crystal Structure of the complex of human mitogen activated protein kinase kinase 5 phox domain (MAP2K5-phox) with human mitogen activated protein kinase kinase kinase 2 phox domain (MAP3K2-phox) | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 5, Mitogen-activated protein kinase kinase kinase 2 | | Authors: | Filippakopoulos, P, Debreczeni, J, Papagrigoriou, V, Turnbull, A, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the complex of human mitogen activated protein kinase kinase 5 phox domain (MAP2K5-phox) with human mitogen activated protein kinase kinase kinase 2 phox domain (MAP3K2-phox)
To be Published
|
|
8F6S
 
 | | LSD1-CoREST in complex with T105 | | Descriptor: | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-(5-methyl-1,3,4-thiadiazol-2-yl)benzamide, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8F30
 
 | | LSD1-CoREST in complex with AW2, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-{5-[3-([1,1'-biphenyl]-4-yl)propanoyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
3SKR
 
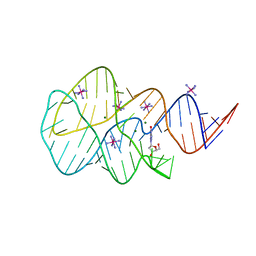 | | Crystal structure of the 2'- Deoxyguanosine riboswitch bound to 2'- Deoxyguanosine, cobalt Hexammine soak | | Descriptor: | 2'-DEOXY-GUANOSINE, COBALT HEXAMMINE(III), MAGNESIUM ION, ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
8F59
 
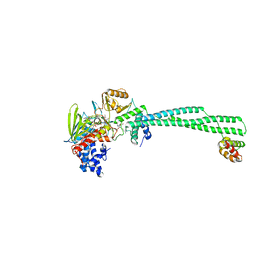 | | LSD1-CoREST in complex with AW2 and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-12 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
3SKZ
 
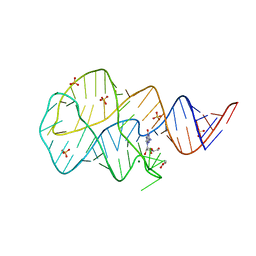 | | Crystal structure of the 2'- deoxyguanosine riboswitch bound to guanosine | | Descriptor: | GUANOSINE, MAGNESIUM ION, RNA (68-MER), ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
8F2Z
 
 | | LSD1-CoREST in complex with AW2, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-([1,1'-biphenyl]-4-yl)-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
3SLZ
 
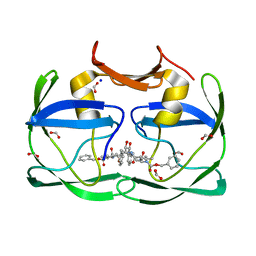 | | The crystal structure of XMRV protease complexed with TL-3 | | Descriptor: | FORMIC ACID, SODIUM ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, ... | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
3SM2
 
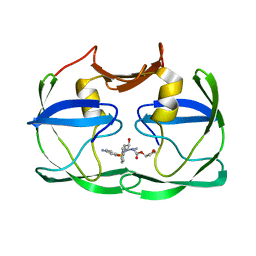 | | The crystal structure of XMRV protease complexed with Amprenavir | | Descriptor: | gag-pro-pol polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
3SLQ
 
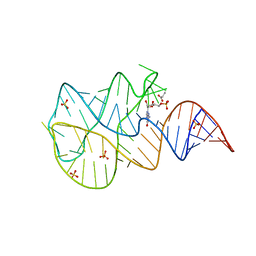 | | Crystal structure of the 2'- Deoxyguanosine riboswitch bound to guanosine-5'-monophosphate | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RNA (68-MER), SUCCINIC ACID, ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-24 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
8FYV
 
 | | Salmonella enterica serovar Typhimurium chemoreceptor Tsr (taxis to serine and repellents) ligand-binding domain in complex with l-serine | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, SERINE, ... | | Authors: | Baylink, A, Gentry-Lear, Z, Glenn, S. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacterial vampirism mediated through taxis to serum.
Elife, 12, 2024
|
|
8FQL
 
 | | Portal vertex of HK97 phage | | Descriptor: | Portal protein | | Authors: | Huet, A, Oh, B, Maurer, J, Duda, R.L, Conway, J.F. | | Deposit date: | 2023-01-06 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A symmetry mismatch unraveled: How phage HK97 scaffold flexibly accommodates a 12-fold pore at a 5-fold viral capsid vertex.
Sci Adv, 9, 2023
|
|
8FMX
 
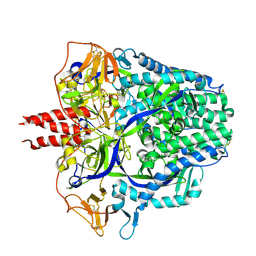 | | Langya virus F glycoprotein ectodomain in prefusion form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Low, Y.S, Isaacs, A, Modhiran, N, Watterson, D. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure and antigenicity of divergent Henipavirus fusion glycoproteins.
Nat Commun, 14, 2023
|
|
