2H42
 
 | | Crystal structure of PDE5 in complex with sildenafil | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
8X1V
 
 | |
8X6V
 
 | | Crystal structure of GlacPETase | | Descriptor: | GlacPETase | | Authors: | Qi, X, Zhou, N.Y. | | Deposit date: | 2023-11-22 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The unique salt bridge network in GlacPETase: a key to its stability.
Appl.Environ.Microbiol., 90, 2024
|
|
2H44
 
 | | Crystal structure of PDE5A1 in complex with icarisid II | | Descriptor: | 5,7-DIHYDROXY-2-(4-METHOXYPHENYL)-8-(3-METHYLBUTYL)-4-OXO-4H-CHROMEN-3-YL 6-DEOXY-ALPHA-L-MANNOPYRANOSIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
2H5E
 
 | | Crystal structure of E.coli polypeptide release factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor RF-3 | | Authors: | Song, H.W, Zhou, Z.H. | | Deposit date: | 2006-05-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
3VZE
 
 | |
8HFE
 
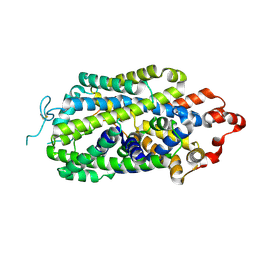 | | Cryo-EM structure of human norepinephrine transporter NET in an inward-open state at resolution of 2.5 angstrom | | Descriptor: | CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
8HFF
 
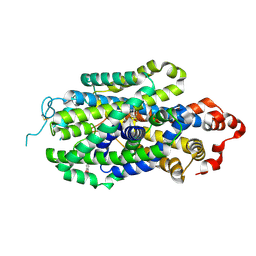 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of norepinephrine in an inward-open state at resolution of 2.9 angstrom. | | Descriptor: | L-NOREPINEPHRINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
8HFL
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant bupropion in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | Bupropion, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
6NT2
 
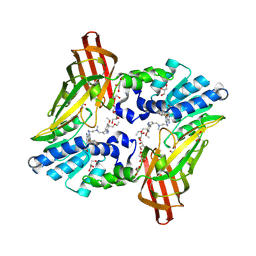 | | type 1 PRMT in complex with the inhibitor GSK3368715 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, N~1~-({5-[4,4-bis(ethoxymethyl)cyclohexyl]-1H-pyrazol-4-yl}methyl)-N~1~,N~2~-dimethylethane-1,2-diamine, ... | | Authors: | Concha, N.O. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss.
Cancer Cell, 36, 2019
|
|
4YDM
 
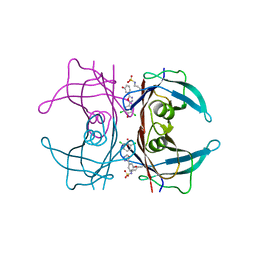 | | High resolution crystal structure of human transthyretin bound to ligand and conjugates of 3-(5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl)phenyl fluorosulfate | | Descriptor: | 2,6-dichloro-4-[5-(3-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]phenol, Transthyretin | | Authors: | Connelly, S, Bradbury, N.C. | | Deposit date: | 2015-02-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Fluorogenic Aryl Fluorosulfate for Intraorganellar Transthyretin Imaging in Living Cells and in Caenorhabditis elegans.
J.Am.Chem.Soc., 137, 2015
|
|
4Y2L
 
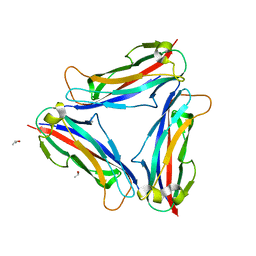 | | Structure of CFA/I pili major subunit CfaB trimer | | Descriptor: | CALCIUM ION, CFA/I fimbrial subunit B, ISOPROPYL ALCOHOL | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2015-02-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Mol. Microbiol., 102, 2016
|
|
4YDN
 
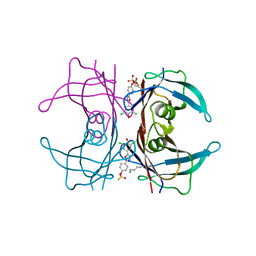 | | High resolution crystal structure of human transthyretin bound to ligand and conjugates of 4-(5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl)phenyl fluorosulfate | | Descriptor: | 2,6-dichloro-4-[5-(4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]phenol, 4-[5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]phenyl sulfurofluoridate, Transthyretin | | Authors: | Connelly, S, Bradbury, N.C. | | Deposit date: | 2015-02-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Fluorogenic Aryl Fluorosulfate for Intraorganellar Transthyretin Imaging in Living Cells and in Caenorhabditis elegans.
J.Am.Chem.Soc., 137, 2015
|
|
3VY6
 
 | |
4Y2O
 
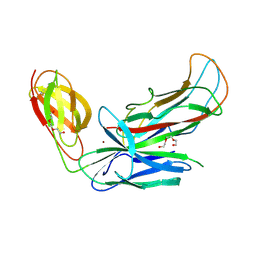 | | Structure of CFA/I pili chaperone-major subunit complex (CfaA-CfaB) | | Descriptor: | CFA/I fimbrial subunit B, CfA/I fimbrial subunit A (Colonization factor antigen subunit A putative chaperone), NICKEL (II) ION, ... | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2015-02-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Mol. Microbiol., 102, 2016
|
|
4XH3
 
 | |
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8J19
 
 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J1A
 
 | | Cryo-EM structure of the GPR84 receptor-Gi complex with no ligand modeled | | Descriptor: | Antibody fragment ScFv16, G-protein coupled receptor 84, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
2EW6
 
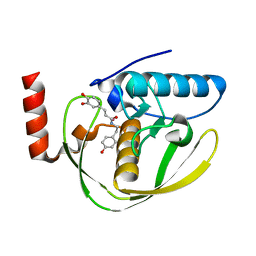 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | (2E)-3-(3,4-DIHYDROXYPHENYL)-N-[2-(4-HYDROXYPHENYL)ETHYL]ACRYLAMIDE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2GYX
 
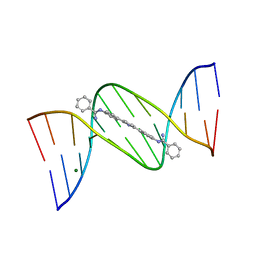 | | Crystal structure of DB884- D(CGCGAATTCGCG)2 complex at 1.86 A resolution. | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-(1H-PYRROLE-2,5-DIYLDI-4,1-PHENYLENE)DIBENZENECARBOXIMIDAMIDE | | Authors: | Lee, M.P.H, Neidle, S. | | Deposit date: | 2006-05-10 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Induced fit conformational changes of a "reversed amidine" heterocycle: optimized interactions in a DNA minor groove complex.
J.Am.Chem.Soc., 129, 2007
|
|
2IIE
 
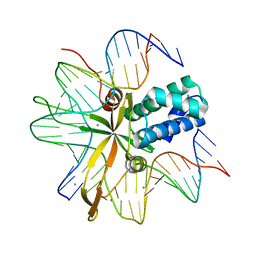 | | single chain Integration Host Factor protein (scIHF2) in complex with DNA | | Descriptor: | DNA (5'-D(*DGP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DAP*DTP*DTP*DTP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DCP*DAP*DAP*DAP*DAP*DAP*DAP*DGP*DCP*DAP*DTP*DT)-3'), Integration host factor, ... | | Authors: | Bao, Q, Droege, P, Davey, C.A. | | Deposit date: | 2006-09-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Divalent Metal-mediated Switch Controlling Protein-induced DNA Bending
J.Mol.Biol., 367, 2007
|
|
