1SQV
 
 | | Crystal Structure Analysis of Bovine Bc1 with UHDBT | | Descriptor: | 6-HYDROXY-5-UNDECYL-1,3-BENZOTHIAZOLE-4,7-DIONE, Cytochrome b, Cytochrome c1, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-09-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
6P6W
 
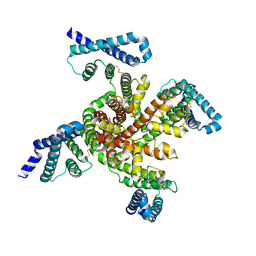 | | Cryo-EM structure of voltage-gated sodium channel NavAb N49K/L109A/M116V/G94C/Q150C disulfide crosslinked mutant in the resting state | | Descriptor: | Fusion of Maltose-binding protein and voltage-gated sodium channel NavAb | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-Din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
5I2Z
 
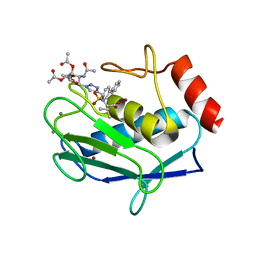 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated triazole-linked carboxylate chelating water-soluble inhibitor (DC24). | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
1SQX
 
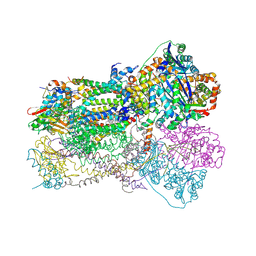 | | Crystal Structure Analysis of Bovine Bc1 with Stigmatellin A | | Descriptor: | Cytochrome b, Cytochrome c1, heme protein, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
5I0L
 
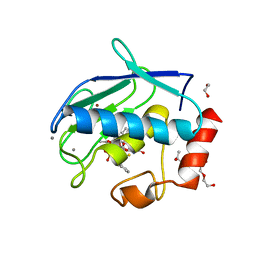 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
5TGL
 
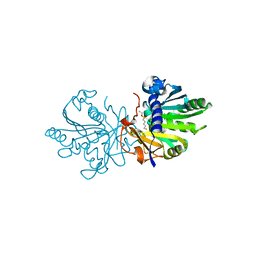 | | A MODEL FOR INTERFACIAL ACTIVATION IN LIPASES FROM THE STRUCTURE OF A FUNGAL LIPASE-INHIBITOR COMPLEX | | Descriptor: | LIPASE, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Brzozowski, A.M, Derewenda, U, Derewenda, Z.S, Dodson, G.G, Lawson, D, Turkenburg, J.P, Bjorkling, F, Huge-Jensen, B, Patkar, S.R, Thim, L. | | Deposit date: | 1991-10-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex.
Nature, 351, 1991
|
|
5XIW
 
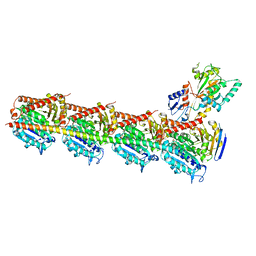 | | Crystal structure of T2R-TTL-Colchicine complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-04-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
6P6Y
 
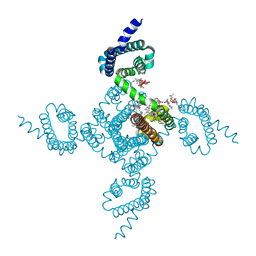 | | Crystal structure of voltage-gated sodium channel NavAb V100C/Q150C disulfide crosslinked mutant in the activated state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
5C9C
 
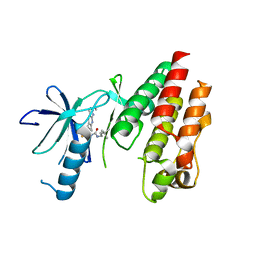 | | CRYSTAL STRUCTURE OF BRAF(V600E) IN COMPLEX WITH LY3009120 COMPND | | Descriptor: | 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Edwards, T, Abendroth, J, Chun, L. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers.
Cancer Cell, 28, 2015
|
|
6P6X
 
 | | Crystal structure of voltage-gated sodium channel NavAb G94C/Q150C mutant in the activated state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-Din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
7T4Q
 
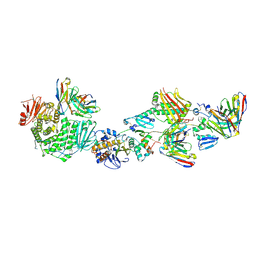 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
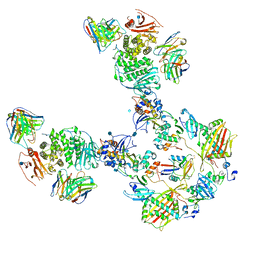 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4S
 
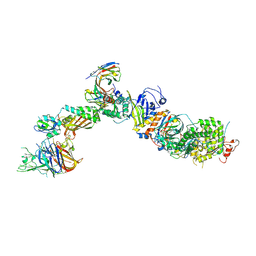 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
5XP3
 
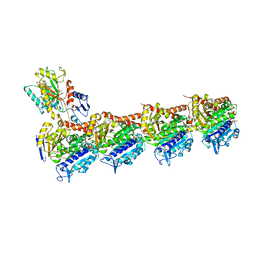 | | Crystal structure of apo T2R-TTL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
6PQ0
 
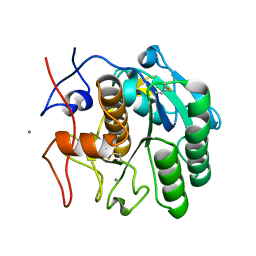 | | LCP-embedded Proteinase K treated with MPD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
6PQ4
 
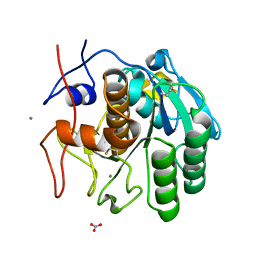 | | LCP-embedded Proteinase K treated with lipase | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | Authors: | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
6PT2
 
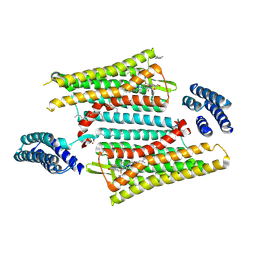 | | Crystal structure of the active delta opioid receptor in complex with the peptide agonist KGCHM07 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Delta opioid receptor, ... | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
5II6
 
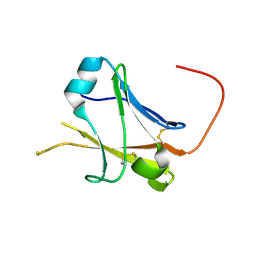 | | Crystal structure of the ZP-N1 domain of mouse sperm receptor ZP2 at 0.95 A resolution | | Descriptor: | Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Han, L, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
6Q8J
 
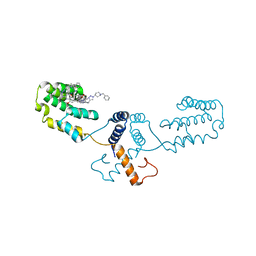 | | Nterminal domain of human SMU1 in complex with LSP641 | | Descriptor: | 1-~{tert}-butyl-3-[6-(3,5-dimethoxyphenyl)-2-[[1-[1-(phenylmethyl)piperidin-4-yl]-1,2,3-triazol-4-yl]methylamino]pyrido[2,3-d]pyrimidin-7-yl]urea, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7O25
 
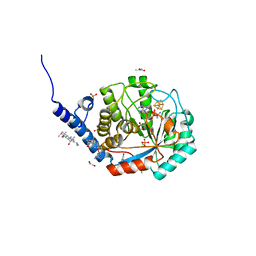 | | Complex-B bound [FeFe]-hydrogenase maturase HydE from T. maritima (reaction triggered in the crystal) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1P
 
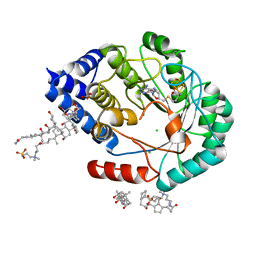 | | [FeFe]-hydrogenase maturase HydE from T. Maritima (C-ter stretch absent) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
