6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4UE5
 
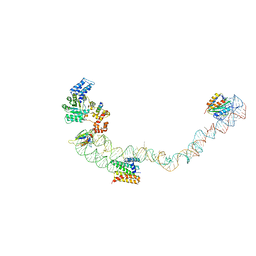 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5Q00
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 2-[3-[(5-bromo-1,3-thiazol-2-yl)carbamoylsulfamoyl]-1-methylindol-7-yl]oxyacetamide | | Descriptor: | 2-[(3-{[(5-bromo-1,3-thiazol-2-yl)carbamoyl]sulfamoyl}-1-methyl-1H-indol-7-yl)oxy]acetamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 2-[3-[(5-bromo-1,3-thiazol-2-yl)carbamoylsulfamoyl]-1-methylindol-7-yl]oxyacetamide
To be published
|
|
4UER
 
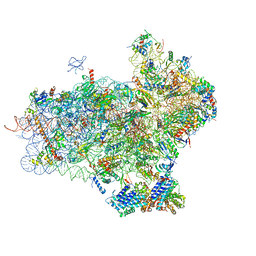 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
6TFY
 
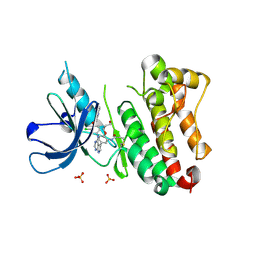 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 18c | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[5-[4-[[3-chloranyl-4-(pyridin-2-ylmethoxy)phenyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]-2-(3-oxidanylpropoxy)phenyl]propanamide | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
6TTV
 
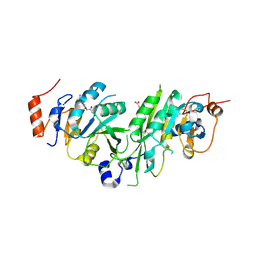 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 3 (ASI_M3M_138) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6THE
 
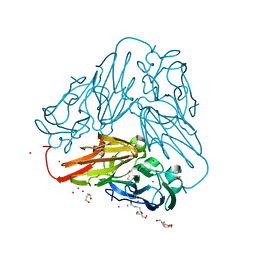 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6TUI
 
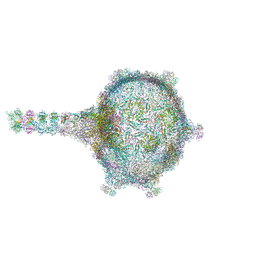 | | Virion of empty GTA particle | | Descriptor: | Adaptor protein Rcc01688, Phage major capsid protein, HK97 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (10.47 Å) | | Cite: | Virion of empty GTA particle
Nat Commun, 11, 2020
|
|
4UFP
 
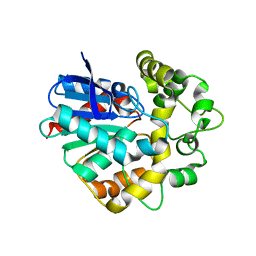 | | Laboratory evolved variant R-C1B1D33 of potato epoxide hydrolase StEH1 | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Carlsson, A.J, Bauer, P, Nilsson, M, Dobritzsch, D, Kamerlin, S.C.L, Widersten, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Laboratory Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis.
Chembiochem, 17, 2016
|
|
3DHA
 
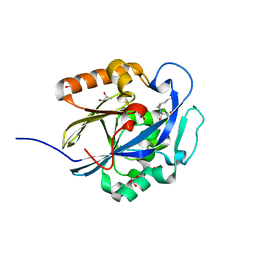 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
6T4H
 
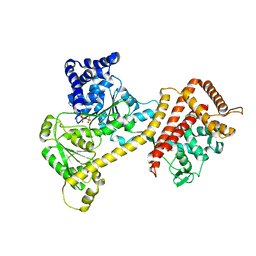 | | Crystal structure of the accessory translocation ATPase, SecA2, from Clostridium difficile, in complex with adenosine-5'-(gamma-thio)-triphosphate | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein translocase subunit SecA 2 | | Authors: | Lindic, N, Loboda, J, Usenik, A, Turk, D. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of Clostridioides difficile SecA2 ATPase Exposes Regions Responsible for Differential Target Recognition of the SecA1 and SecA2-Dependent Systems.
Int J Mol Sci, 21, 2020
|
|
6TM5
 
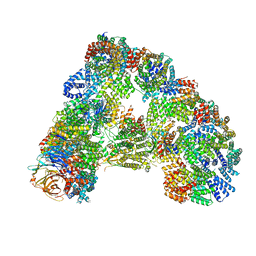 | |
4UEE
 
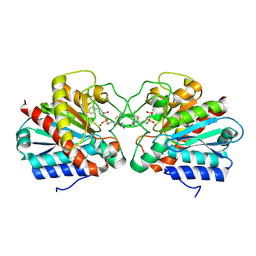 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with the PHOSPHINIC INHIBITOR Acetyl-Leu-Ala-Y(PO2CH2)-homoPhe-OH | | Descriptor: | (2S)-2-{[(R)-{(1R)-1-[(N-acetyl-L-leucyl)amino]ethyl}(hydroxy)phosphoryl]methyl}-4-phenylbutanoic acid, CARBOXYPEPTIDASE A1, ZINC ION | | Authors: | Gallego, P, Covaleda, G, Costenaro, L, Devel, L, Dive, V, Aviles, F.X, Reverter, D. | | Deposit date: | 2014-12-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Carboxypeptidase A1 in Complex with Phosphinic Inhibitors
To be Published
|
|
5PZY
 
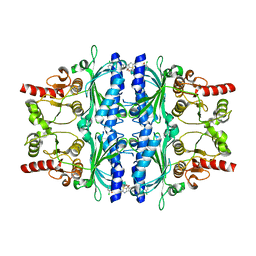 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-(2-chlorophenyl)sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-2-chlorobenzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-(2-chlorophenyl)sulfonylurea
To be published
|
|
4UCV
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
6TFO
 
 | | Crystal structure of as isolated three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
6TS9
 
 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-lactamase, ... | | Authors: | Maso, L, Tondi, D, Klein, R, Montanari, M, Bellio, C, Celenza, G, Brenk, R, Cendron, L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the Class A Carbapenemase GES-5 via Virtual Screening.
Biomolecules, 10, 2020
|
|
6TG8
 
 | | Crystal structure of the Kelch domain in complex with 11 amino acid peptide (model of the ETGE loop) | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, VAL-ILE-ASN-PRO-GLU-THR-GLY-GLU-GLN-ILE-GLN | | Authors: | Kekez, I, Matic, S, Tomic, S, Matkovic-Calogovic, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Binding of dipeptidyl peptidase III to the oxidative stress cell sensor Kelch-like ECH-associated protein 1 is a two-step process.
J.Biomol.Struct.Dyn., 39, 2021
|
|
4UI9
 
 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
6T2W
 
 | | Crystal structure of the CSF1R kinase domain with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Macrophage colony-stimulating factor 1 receptor, SULFATE ION | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
4UHB
 
 | | Laboratory evolved variant R-C1 of potato epoxide hydrolase StEH1 | | Descriptor: | 1,2-ETHANEDIOL, EPOXIDE HYDROLASE, GLYCEROL | | Authors: | Nilsson, M.T.I, Carlsson, A.J, Dobritzsch, D, Widersten, M. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Laboratory Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis.
Chembiochem, 17, 2016
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
5PZT
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-(3-ethyl-4-phenylphenyl)sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-2-ethyl[1,1'-biphenyl]-4-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-(3-ethyl-4-phenylphenyl)sulfonylurea
To be published
|
|
6TE2
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the 2-aminothiazole-type inhibitor 17 | | Descriptor: | 3-[(4-pyridin-2-yl-1,3-thiazol-2-yl)amino]benzoic acid, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.922 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
