3JT0
 
 | | Crystal Structure of the C-terminal fragment (426-558) Lamin-B1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR5546A | | Descriptor: | Lamin-B1 | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-11 | | Release date: | 2009-09-22 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR5546A
To be Published
|
|
3K20
 
 | | X-ray structure of oxidoreductase from corynebacterium diphtheriae,hexagonal crystal form. northeast structural genomics consortium target cdr100d | | Descriptor: | SULFATE ION, oxidoreductase | | Authors: | Kuzin, A, Lew, S, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-20 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target CdR100D
To be Published
|
|
3K94
 
 | | Crystal Structure of Thiamin pyrophosphokinase from Geobacillus thermodenitrificans, Northeast Structural Genomics Consortium Target GtR2 | | Descriptor: | Thiamin pyrophosphokinase | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Janjua, J, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Northeast Structural Genomics Consortium Target GtR2
To be Published
|
|
6WAT
 
 | | complex structure of PHF1 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 1, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
3KB1
 
 | | Crystal Structure of the Nucleotide-binding protein AF_226 in complex with ADP from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR157 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleotide-binding protein, ZINC ION | | Authors: | Forouhar, F, Lew, S, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR157
To be Published
|
|
3K1Y
 
 | | X-ray structure of oxidoreductase from corynebacterium diphtheriae. orthorombic crystal form, northeast structural genomics consortium target cdr100d | | Descriptor: | SULFATE ION, oxidoreductase | | Authors: | Kuzin, A, Lew, S, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-20 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target CdR100D
To be Published
|
|
8QWL
 
 | | Structure of p53 cancer mutant Y163C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, MALONATE ION, ... | | Authors: | Balourdas, D.I, Markl, A.M, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWM
 
 | | Structure of p53 cancer mutant Y205C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWN
 
 | | Structure of p53 cancer mutant Y220C (hexagonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
3JVN
 
 | | Crystal Structure of the acetyltransferase VF_1542 from Vibrio fischeri, Northeast Structural Genomics Consortium Target VfR136 | | Descriptor: | Acetyltransferase, SULFATE ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-17 | | Release date: | 2009-09-29 | | Last modified: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Northeast Structural Genomics Consortium Target VfR136
To be Published
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4DRT
 
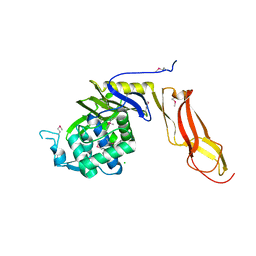 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
8QWK
 
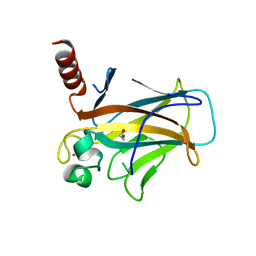 | | Structure of p53 cancer mutant Y126C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Markl, A.M, Balourdas, D.I, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWO
 
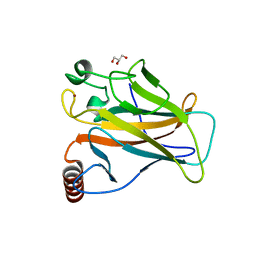 | | Structure of p53 cancer mutant Y234C | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWP
 
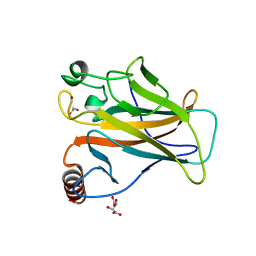 | | Structure of p53 cancer mutant Y236C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
3HVW
 
 | | Crystal Structure of the GGDEF domain of the PA2567 protein from Pseudomonas aeruginosa, Northeast Structural Genomics Consortium Target PaR365C | | Descriptor: | Diguanylate-cyclase (DGC) | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-23 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target PaR365C
To be published
|
|
3HXP
 
 | | Crystal structure of the FhuD fold-family BSU3320, a periplasmic binding protein component of a Fep/Fec-like ferrichrome ABC transporter from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR577 | | Descriptor: | Iron(3+)-hydroxamate-binding protein fhuD | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-21 | | Release date: | 2009-07-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR577
To be Published
|
|
4NZG
 
 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Integrase p46, ... | | Authors: | Guan, R, Jiang, M, Janjua, H, Maglaqui, M, Zhao, L, Xiao, R, Acton, T.B, Everett, J.K, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
6WAU
 
 | | Complex structure of PHF19 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 19, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6WAV
 
 | | Crystal structure of PHF1 in complex with H3K36me3 substitution | | Descriptor: | Histone H3.1, PHD finger protein 1, SULFATE ION, ... | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6DDH
 
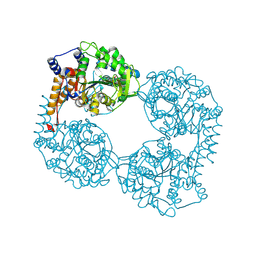 | | Crystal structure of the double mutant (D52N/R367Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, INOSINIC ACID | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
3LRX
 
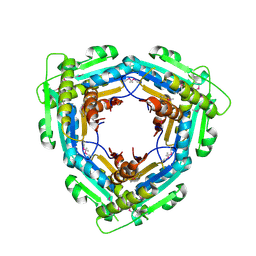 | | Crystal Structure of the C-terminal domain (residues 78-226) of PF1911 hydrogenase from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR246A | | Descriptor: | Putative hydrogenase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Northeast Structural Genomics Consortium Target PfR246A
To be Published
|
|
2AJA
 
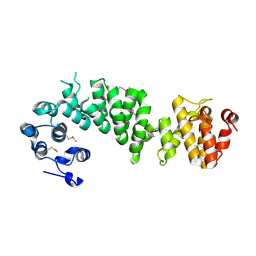 | | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR21. | | Descriptor: | ankyrin repeat family protein | | Authors: | Kuzin, A.P, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, C, Kellie, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila.
To be Published
|
|
4YY8
 
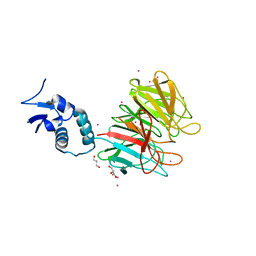 | | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum | | Descriptor: | GLYCEROL, Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum.
to be published
|
|
3LKD
 
 | | Crystal Structure of the type I restriction-modification system methyltransferase subunit from Streptococcus thermophilus, Northeast Structural Genomics Consortium Target SuR80 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, M, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Northeast Structural Genomics Consortium Target SuR80
To be published
|
|
