7EY5
 
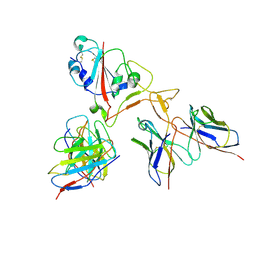 | |
7TP3
 
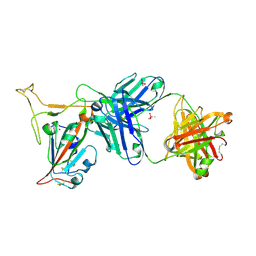 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
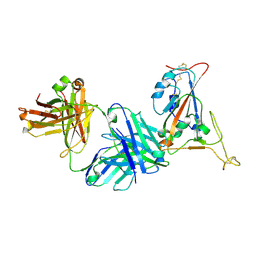 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TLX
 
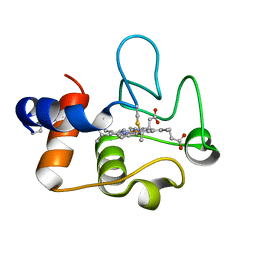 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
7U6L
 
 | | Pseudooxynicotine amine oxidase | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Choudhary, V, Stull, F, Wu, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
7U5K
 
 | | Cryo-EM Structure of DPYSL2 | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7YFY
 
 | | Cryo-EM structure of the Mili-piRNA- target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (5'-R(P*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7U0R
 
 | | Crystal structure of Methanomethylophilus alvus PylRS(N166A/V168A) complexed with meta-trifluoromethyl-2-benzylmalonate and AMP-PNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Swenson, C.V, Roe, L.T, Fricke, R.C.B, Smaga, S.S, Gee, C.L, Schepartz, A. | | Deposit date: | 2022-02-18 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-alpha-amino acids in vitro and in vivo.
Nat.Chem., 15, 2023
|
|
7UG2
 
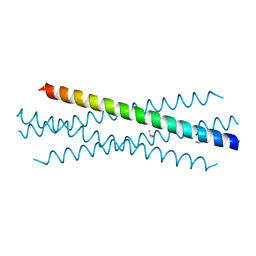 | | Crystal structure of the coiled-coil domain of TRIM75 | | Descriptor: | ACETYL GROUP, ISOPROPYL ALCOHOL, Tripartite motif-containing protein 75 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural studies of the coiled-coil domain of TRIM75 reveal a tetramer architecture facilitating its E3 ligase complex.
Comput Struct Biotechnol J, 20, 2022
|
|
7VTQ
 
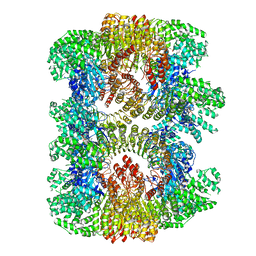 | | Cryo-EM structure of mouse NLRP3 (full-length) dodecamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2021-10-30 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VTP
 
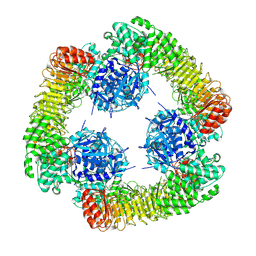 | | Cryo-EM structure of PYD-deleted human NLRP3 hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2021-10-30 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VGI
 
 | |
7YG6
 
 | | Cryo-EM structure of the EfPiwi(N959K) in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YGN
 
 | | Cryo-EM structure of the Mili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFX
 
 | | Cryo-EM structure of Hili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YFQ
 
 | | Cryo-EM structure of the EfPiwi (N959K)-piRNA-target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi, RNA (5'-R(*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7VGH
 
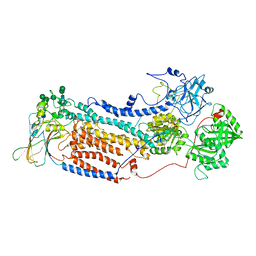 | |
7VGJ
 
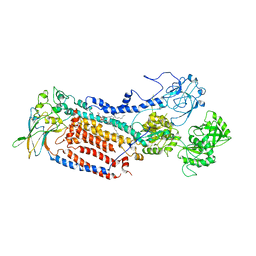 | | Cryo-EM structure of the human P4-type flippase ATP8B1-CDC50A in the auto-inhibited E2Pi-PS state | | Descriptor: | Cell cycle control protein 50A, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Phospholipid-transporting ATPase IC | | Authors: | Chen, M.T, Chen, Y, Chen, Z.P, Zhou, C.Z, Hou, W.T, Chen, Y. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural insights into the activation of autoinhibited human lipid flippase ATP8B1 upon substrate binding.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3A
 
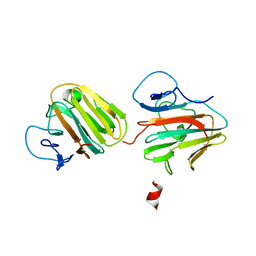 | | Crystal structure of TRIM7 bound to 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3B
 
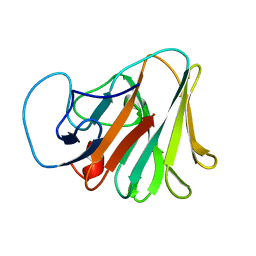 | | Crystal structure of TRIM7 bound to GN1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3C
 
 | | Crystal structure of TRIM7 bound to RACO-1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8T6B
 
 | | Human VMAT2 in complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T69
 
 | | Human VMAT2 in complex with tetrabenazine | | Descriptor: | Synaptic vesicular amine transporter, tetrabenazine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T6A
 
 | | Human VMAT2 in complex with reserpine | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
7DMN
 
 | | Crystal structure of two pericyclases catalyzing [4+2] cycloaddition | | Descriptor: | Diels-Alderase fsa2, GLYCEROL | | Authors: | Chi, C.B, Wang, Z.D, Liu, T, Zhang, Z.Y, Ma, M. | | Deposit date: | 2020-12-04 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Fsa2 and Phm7 Catalyzing [4 + 2] Cycloaddition Reactions with Reverse Stereoselectivities in Equisetin and Phomasetin Biosynthesis.
Acs Omega, 6, 2021
|
|
