6I5N
 
 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with growth hormone receptor peptide | | Descriptor: | COBALT (II) ION, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-14 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
6I5J
 
 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with growth hormone receptor peptide | | Descriptor: | COBALT (II) ION, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
7Z0Y
 
 | | THSC20.HVTR04 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
7Z0X
 
 | | THSC20.HVTR26 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
6EZ9
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor JHU3372 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]oxycarbonylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
6ETY
 
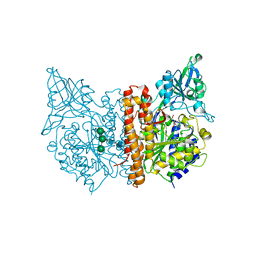 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor JHU3371 | | Descriptor: | (2~{S})-2-[[(2~{R})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoyloxy]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-10-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
6FE5
 
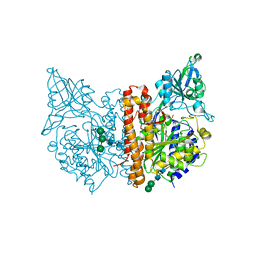 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor JHU 2249 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
6F5L
 
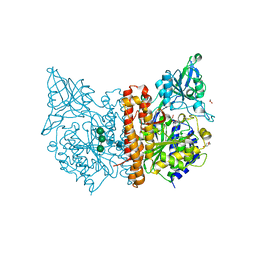 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor JHU2379 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoyloxy]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-12-01 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
5ELY
 
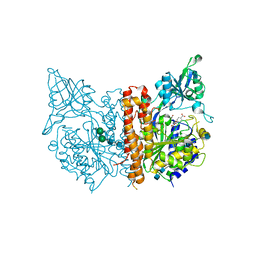 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a hydroxamate inhibitor JHU242 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{R})-2-carboxy-5-(oxidanylamino)-5-oxidanylidene-pentyl]benzoic acid, ... | | Authors: | Barinka, C, Novakova, Z, Pavlicek, J. | | Deposit date: | 2015-11-05 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Unprecedented Binding Mode of Hydroxamate-Based Inhibitors of Glutamate Carboxypeptidase II: Structural Characterization and Biological Activity.
J.Med.Chem., 59, 2016
|
|
5D29
 
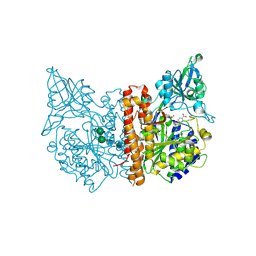 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a hydroxamate inhibitor JHU241 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{S})-2-carboxy-5-(oxidanylamino)-5-oxidanylidene-pentyl]benzoic acid, ... | | Authors: | Barinka, C, Novakova, Z, Pavlicek, J. | | Deposit date: | 2015-08-05 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented Binding Mode of Hydroxamate-Based Inhibitors of Glutamate Carboxypeptidase II: Structural Characterization and Biological Activity.
J.Med.Chem., 59, 2016
|
|
3BI0
 
 | |
3BI1
 
 | |
3BHX
 
 | |
3AQE
 
 | | Crystal structure of the extracellular domain of human RAMP2 | | Descriptor: | Receptor activity-modifying protein 2 | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Protein Sci., 21, 2012
|
|
6ZGM
 
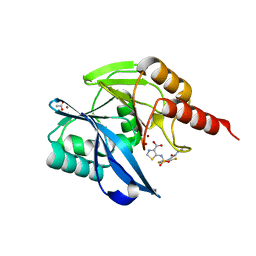 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with the thiazolecarboxylate inhibitor ANT2681 | | Descriptor: | 5-[[4-(carbamimidamidocarbamoylamino)-3,5-bis(fluoranyl)phenyl]sulfonylamino]-1,3-thiazole-4-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Docquier, J.D, Pozzi, C, Marcoccia, F, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ANT2681: SAR Studies Leading to the Identification of a Metallo-beta-lactamase Inhibitor with Potential for Clinical Use in Combination with Meropenem for the Treatment of Infections Caused by NDM-ProducingEnterobacteriaceae.
Acs Infect Dis., 6, 2020
|
|
3AM6
 
 | |
1TRA
 
 | | RESTRAINED REFINEMENT OF THE MONOCLINIC FORM OF YEAST PHENYLALANINE TRANSFER RNA. TEMPERATURE FACTORS AND DYNAMICS, COORDINATED WATERS, AND BASE-PAIR PROPELLER TWIST ANGLES | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Westhof, E, Sundaralingam, M. | | Deposit date: | 1986-05-16 | | Release date: | 1986-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles.
Biochemistry, 25, 1986
|
|
1TYK
 
 | | SOLUTION STRUCTURE OF A TOXIN FROM THE TARANTULA, GRAMMOSTOLA SPATULATA, WHICH INHIBITS MECHANOSENSITIVE ION CHANNELS | | Descriptor: | Toxin GsMTx-4 | | Authors: | Oswald, R.E, Suchyna, T.M, Mcfeeters, R, Gottlieb, P, Sachs, F. | | Deposit date: | 2004-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Peptide Toxins that Block Mechanosensitive Ion Channels
J.Biol.Chem., 277, 2002
|
|
2DRX
 
 | | Structure Analysis of (POG)4-(LOG)2-(POG)4 | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
2DRT
 
 | | Structure Analysis of (POG)4-LOG-(POG)5 | | Descriptor: | ETHANOL, collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
6XOG
 
 | | Structure of SUMO1-ML786519 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
3AOI
 
 | | RNA polymerase-Gfh1 complex (Crystal type 2) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*A*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
3AOH
 
 | | RNA polymerase-Gfh1 complex (Crystal type 1) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*AP*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
