6C16
 
 | |
6BVA
 
 | |
6CPM
 
 | | Structure of the USP15 deubiquitinase domain in complex with a third-generation inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6CRN
 
 | | Structure of the USP15 deubiquitinase domain in complex with a high-affinity first-generation Ubv | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 15, Ubiquitin variant 15.2, ZINC ION | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6BYH
 
 | |
6CAD
 
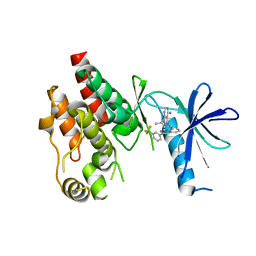 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
6BXI
 
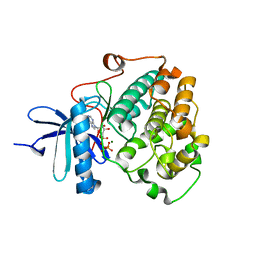 | | X-ray crystal structure of NDR1 kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase 38 | | Authors: | Xiong, S, Sicheri, F. | | Deposit date: | 2017-12-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Auto-Inhibition of the NDR1 Kinase Domain by an Atypically Long Activation Segment.
Structure, 26, 2018
|
|
6D4P
 
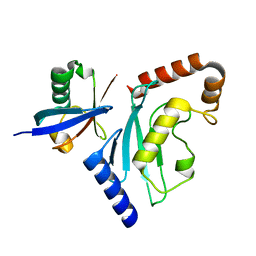 | | Ube2D1 in complex with ubiquitin variant Ubv.D1.1 | | Descriptor: | Ubiquitin Variant Ubv.D1.1, Ubiquitin-conjugating enzyme E2 D1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D68
 
 | | Ube2G1 in complex with ubiquitin variant Ubv.G1.1 | | Descriptor: | Ubiquitin-conjugating enzyme E2 G1, Ubv.G1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-20 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6DGF
 
 | | Ubiquitin Variant bound to USP2 | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 2, ... | | Authors: | Manczyk, N, Sicheri, F. | | Deposit date: | 2018-05-17 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Yeast Two-Hybrid Analysis for Ubiquitin Variant Inhibitors of Human Deubiquitinases.
J. Mol. Biol., 431, 2019
|
|
6D6I
 
 | | Ube2V1 in complex with ubiquitin variant Ubv.V1.1 and Ube2N/Ubc13 | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1, Ubv.V1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Keszei, A, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
7T1K
 
 | | Crystal structure of a superbinder Fes SH2 domain (sFes1) in complex with a high affinity phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1L
 
 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | Descriptor: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1U
 
 | | Crystal structure of a superbinder Src SH2 domain (sSrcF) in complex with a high affinity phosphopeptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, Synthetic phosphopeptide, ZINC ION | | Authors: | Martyn, G.D, Singer, A.U, Manczyk, N, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
2P0F
 
 | | ArhGAP9 PH domain in complex with Ins(1,3,5)P3 | | Descriptor: | PHOSPHATE ION, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
2P0H
 
 | | ArhGAP9 PH domain in complex with Ins(1,3,4)P3 | | Descriptor: | (1S,3S,4S)-1,3,4-TRIPHOSPHO-MYO-INOSITOL, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
2P0D
 
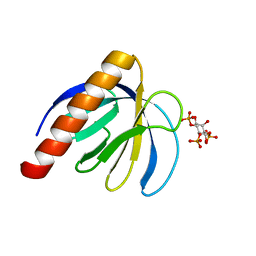 | | ArhGAP9 PH domain in complex with Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
2QNJ
 
 | |
2PBC
 
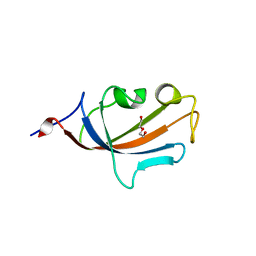 | | FK506-binding protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FK506-binding protein 2 | | Authors: | Walker, J.R, Neculai, D, Davis, T, Butler-Cole, C, Sicheri, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of FK506-Binding Protein 2
To be Published
|
|
3TWQ
 
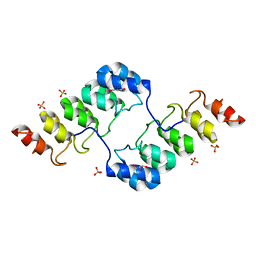 | |
3TWW
 
 | |
3TWS
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human TERF1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TWV
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human NUMA1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TWX
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human FNBP1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TZM
 
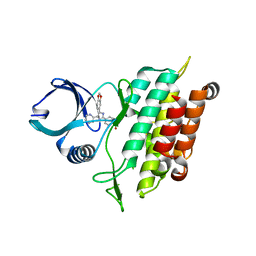 | | TGF-beta Receptor type 1 in complex with SB431542 | | Descriptor: | 4-[5-(1,3-benzodioxol-5-yl)-4-(pyridin-2-yl)-1H-imidazol-2-yl]benzamide, TGF-beta receptor type-1 | | Authors: | Ogunjimi, A.A, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Specificity of TGFbeta Family Receptor Small Molecule Inhibitors
Cell Signal, 24, 2012
|
|
