8C2H
 
 | | Transmembrane domain of active state homomeric GluA1 AMPA receptor in tandem with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1P
 
 | | Active state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CYCLOTHIAZIDE, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1S
 
 | | Transmembrane domain of resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C2I
 
 | | Transmembrane domain of resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1R
 
 | | Resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma-2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1Q
 
 | | Resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8BZS
 
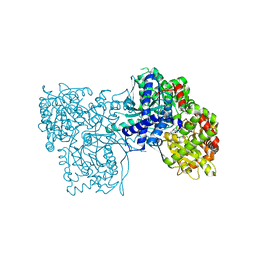 | | The crystal structure of glycogen phosphorylase in complex with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Koulas, S.M, Mathomes, R, Hayes, J.M, Leonidas, D.D. | | Deposit date: | 2022-12-15 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Multidisciplinary docking, kinetics and X-ray crystallography studies of baicalein acting as a glycogen phosphorylase inhibitor and determination of its' potential against glioblastoma in cellular models.
Chem.Biol.Interact., 382, 2023
|
|
8CHB
 
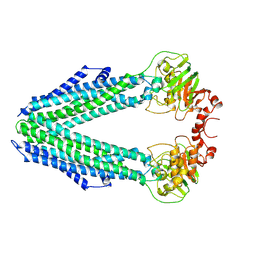 | | Inward-facing conformation of the ABC transporter BmrA C436S/A582C cross-linked mutant | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Hanssen, E, Valimehr, S, Orelle, C, Jault, J.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
5VWZ
 
 | | Bak in complex with Bim-h3Pc | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, AMMONIUM ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.622 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5VZS
 
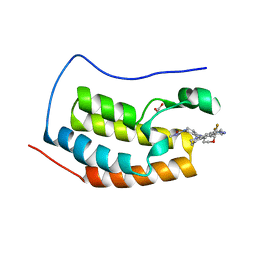 | | BRD4-BD1 in complex with Cpd19 (3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, Bromodomain-containing protein 4 | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-29 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of CBP/P300
To be published
|
|
5VWY
 
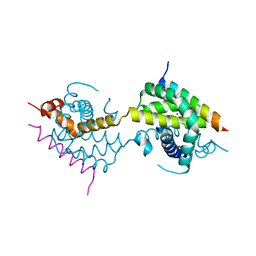 | |
5VX3
 
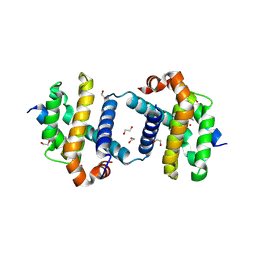 | | Bcl-xL in complex with Bim-h3Pc-RT | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein 1, Bcl-2-like protein 11 | | Authors: | Cowan, A.D, Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5VZB
 
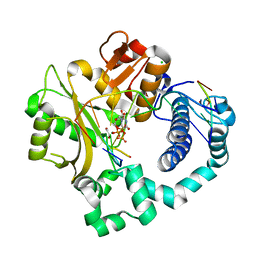 | | Post-catalytic complex of human Polymerase Mu (G433S) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5VZH
 
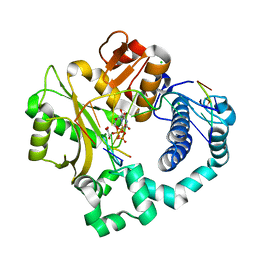 | | Post-catalytic complex of human Polymerase Mu (W434H) mutant with incoming UTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5VRH
 
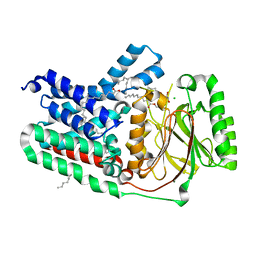 | | Apolipoprotein N-acyltransferase C387S active site mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, ... | | Authors: | Murray, J.M, Noland, C.L. | | Deposit date: | 2017-05-10 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VWX
 
 | | Bak core latch dimer in complex with Bim-h0-h3Glt | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 11 | | Authors: | Brouwer, J.M, Lan, P, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5VZ9
 
 | | Post-catalytic complex of human Polymerase Mu (G433A) mutant with incoming dTTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5VZG
 
 | | Pre-catalytic ternary complex of human Polymerase Mu (W434H) mutant with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5W89
 
 | | Crystal structure of human Mcl-1 in complex with modified Bim BH3 peptide SAH-MS1-18 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, modified Bim BH3 peptide SAH-MS1-18 | | Authors: | Rezaei Araghi, R, Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W90
 
 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
5W8F
 
 | | Crystal structure of human Mcl-1 in complex with modified Bim BH3 peptide SAH-MS1-14 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, modified Bim BH3 peptide SAH-MS1-14 | | Authors: | Rezaei Araghi, R, Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | Descriptor: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | Authors: | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
5WCK
 
 | | Native FEZ-1 metallo-beta-lactamase from Legionella gormanii | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Papamicael, C, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
1MLX
 
 | | Crystal Structure Analysis of a 2'-O-[2-(Methylthio)-ethyl]-Modified Oligodeoxynucleotide Duplex | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*SMTP*AP*CP*GP*C)-3' | | Authors: | Prakash, T.P, Manoharan, M, Kawasaki, A.M, Fraser, A.S, Lesnik, E.A, Sioufi, N, Leeds, J.M, Teplova, M, Egli, M. | | Deposit date: | 2002-08-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 2'-O-[2-(Methylthio)ethyl]-Modified Oligonucleotide: An Analogue of 2'-O-[2-(Methoxy)-ethyl]-Modified
Oligonucleotide with Improved Protein Binding Properties and High Binding Affinity to Target RNA
Biochemistry, 41, 2002
|
|
6R3P
 
 | |
