5YEH
 
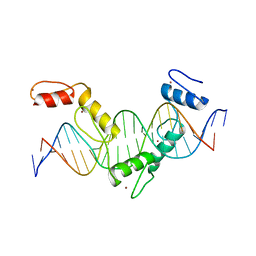 | | Crystal structure of CTCF ZFs4-8-eCBS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5YEF
 
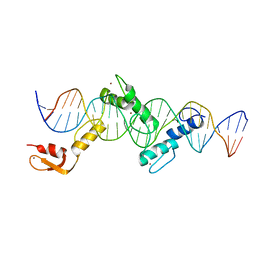 | | Crystal structure of CTCF ZFs2-8-Hs5-1aE | | Descriptor: | DNA (27-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5E1E
 
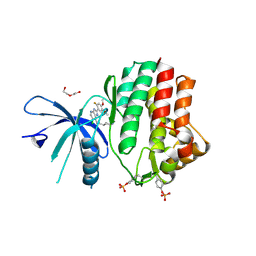 | | Human JAK1 kinase in complex with compound 30 at 2.30 Angstroms resolution | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, DI(HYDROXYETHYL)ETHER, Tyrosine-protein kinase JAK1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2015-12-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of azabenzimidazoles as potent JAK1 selective inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8JGG
 
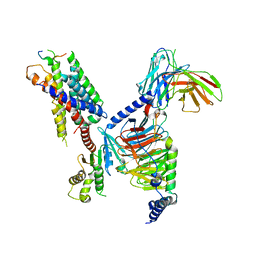 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Ynag, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
8JGB
 
 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist CNF-Tx2 | | Descriptor: | Conorfamide-Tx2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
6JTD
 
 | | Crystal structure of TcCGT1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, C-glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhao, P, Yun, C.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular and Structural Characterization of a Promiscuous C-Glycosyltransferase from Trollius chinensis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6U63
 
 | | Mcl-1 bound to compound 17 | | Descriptor: | 2-{[(naphthalen-2-yl)sulfonyl]amino}-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
7CH7
 
 | | Cryo-EM structure of E.coli MlaFEB | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, Phospholipid ABC transporter ATP-binding protein MlaF | | Authors: | Zhou, C, Shi, H, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
6U6F
 
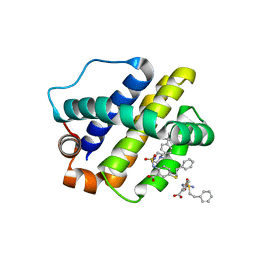 | | The crystal structure of anti-apoptotic Mcl-1 protein in complex with 2, 5-substituted benzoic acid inhibitor 21 | | Descriptor: | 2-[({4-[(4-tert-butylphenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Yang, Y, Stuckey, J.A, Nikolovska-Coleska, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6A58
 
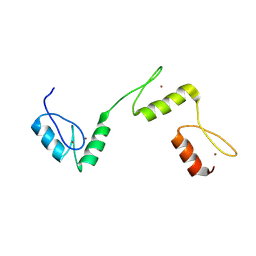 | | Structure of histone demethylase REF6 | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
6Z3S
 
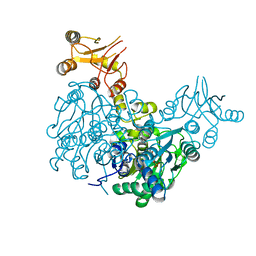 | | Crystal structure of delta466-491 cystathionine beta-synthase from Toxoplasma gondii with O-Acetylserine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine beta-synthase | | Authors: | Fernandez-Rodriguez, C, Oyenarte, I, Conter, C, Gonzalez-Recio, I, Quintana, I, Martinez-Chantar, M, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2020-05-21 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.048 Å) | | Cite: | Structural insight into the unique conformation of cystathionine beta-synthase from Toxoplasma gondii .
Comput Struct Biotechnol J, 19, 2021
|
|
6U65
 
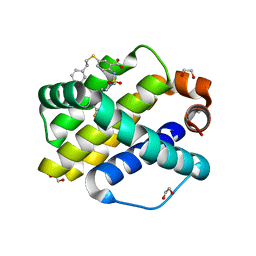 | | Mcl-1 bound to compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 2-[({4-[(4-fluorophenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, ACETATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
8K3D
 
 | | Crystal structure of NRF1 DBD bound to DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*CP*GP*CP*AP*TP*GP*CP*GP*CP*AP*CP*C)-3'), Nuclear respiratory factor 1 | | Authors: | Li, W.F, Liu, K, Min, J.R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of specific DNA sequence recognition by NRF1.
Nucleic Acids Res., 52, 2024
|
|
8K4L
 
 | |
6U64
 
 | | Mcl-1 bound to compound 17 | | Descriptor: | 5-[(2-phenylethyl)sulfanyl]-2-{[(4-phenylpiperazin-1-yl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6U67
 
 | | Mcl-1 bound to compound 24 | | Descriptor: | 2-({[4-(4-tert-butylphenyl)piperazin-1-yl]sulfonyl}amino)-5-{[3-oxo-3-(phenylamino)propyl]sulfanyl}benzoic acid, BIPHENYL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
5X41
 
 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
6A59
 
 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
5X3X
 
 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
6ZS7
 
 | | Crystal structure of delta466-491 cystathionine beta-synthase from Toxoplasma gondii with L-cysteine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine beta-synthase | | Authors: | Fernandez-Rodriguez, C, Oyenarte, I, Conter, C, Gonzalez-Recio, I, Quintana, I, Martinez-Chantar, M, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the unique conformation of cystathionine beta-synthase from Toxoplasma gondii .
Comput Struct Biotechnol J, 19, 2021
|
|
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
5TIO
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
8JGF
 
 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
