6V3N
 
 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V8W
 
 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6ILP
 
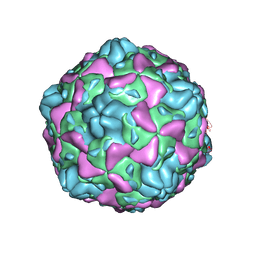 | | Cryo-EM structure of full Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILN
 
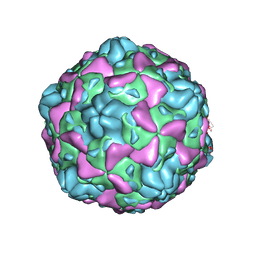 | | Cryo-EM structure of full Echovirus 6 particle at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILK
 
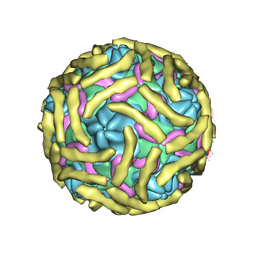 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
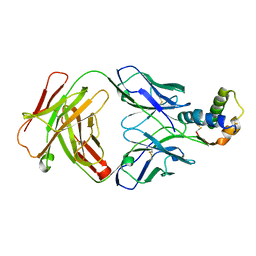
6S3D
 
 | |
6S8B
 
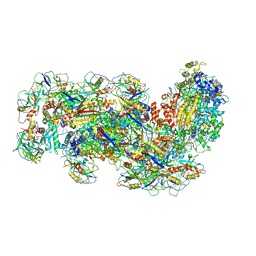 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S91
 
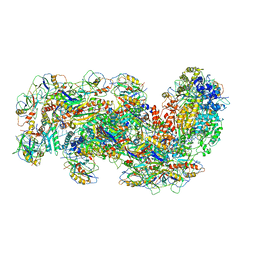 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
3JSG
 
 | |
6S8E
 
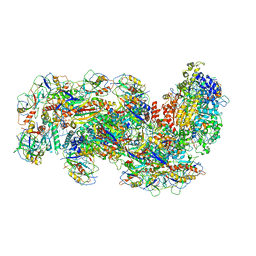 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SH8
 
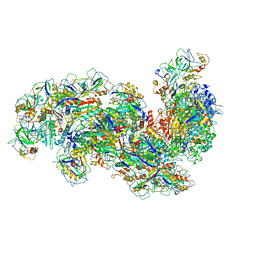 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
3JTU
 
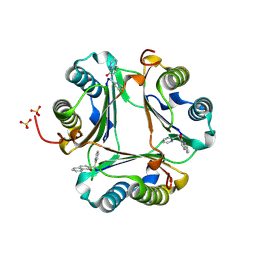 | |
6SHB
 
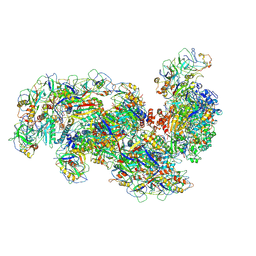 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
3JSF
 
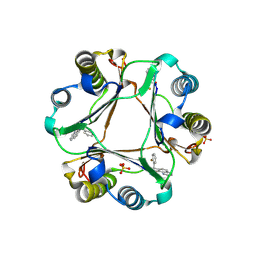 | |
6S6B
 
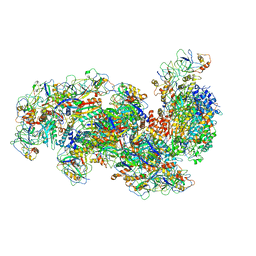 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
7VLN
 
 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | Descriptor: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | Authors: | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
6SIC
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
2MXB
 
 | |
1K94
 
 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K95
 
 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4FVD
 
 | | Crystal structure of EV71 2A proteinase C110A mutant in complex with substrate | | Descriptor: | 10-mer peptide from 2A proteinase, 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
4FVB
 
 | | Crystal structure of EV71 2A proteinase C110A mutant | | Descriptor: | 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
