7N4S
 
 | | Bruton's tyrosine kinase in complex with compound 65 | | Descriptor: | (3R,3'R)-3-anilino-1'-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)[1,3'-bipiperidin]-2-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-06-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Utilizing structure based drug design and metabolic soft spot identification to optimize the in vitro potency and in vivo pharmacokinetic properties leading to the discovery of novel reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 44, 2021
|
|
7N4R
 
 | | Bruton's tyrosine kinase in complex with compound 21 | | Descriptor: | DIMETHYL SULFOXIDE, N-{2-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]ethyl}-N~2~-phenylglycinamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-06-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Utilizing structure based drug design and metabolic soft spot identification to optimize the in vitro potency and in vivo pharmacokinetic properties leading to the discovery of novel reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 44, 2021
|
|
7N4Q
 
 | | Bruton's tyrosine kinase in complex with compound 45 | | Descriptor: | (2R)-2-(3-chloro-5-fluoroanilino)-2-cyclopropyl-N-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]acetamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-06-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Utilizing structure based drug design and metabolic soft spot identification to optimize the in vitro potency and in vivo pharmacokinetic properties leading to the discovery of novel reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 44, 2021
|
|
1P5Q
 
 | | Crystal Structure of FKBP52 C-terminal Domain | | Descriptor: | FK506-binding protein 4, SULFATE ION | | Authors: | Wu, B, Li, P, Lou, Z, Shu, C, Ding, Y, Shen, B, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
9CSY
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to PF-07957472 | | Descriptor: | 2-methyl-5-(4-methylpiperazin-1-yl)-N-{1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]cyclopropyl}benzamide, Papain-like protease, ZINC ION, ... | | Authors: | Mashalidis, E.H, Chang, J.S, Wu, H, Garnsey, M. | | Deposit date: | 2024-07-24 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Discovery of SARS-CoV-2 papain-like protease (PL pro ) inhibitors with efficacy in a murine infection model.
Sci Adv, 10, 2024
|
|
6RYK
 
 | | Crystal structure of the ParB-like protein PadC | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2019-06-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ParB-type DNA Segregation Proteins Are CTP-Dependent Molecular Switches.
Cell, 179, 2019
|
|
3O7P
 
 | | Crystal structure of the E.coli Fucose:proton symporter, FucP (N162A) | | Descriptor: | L-fucose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Dang, S.Y, Sun, L.F, Wang, J, Yan, N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of a fucose transporter in an outward-open conformation
Nature, 467, 2010
|
|
3O7Q
 
 | | Crystal structure of a Major Facilitator Superfamily (MFS) transporter, FucP, in the outward conformation | | Descriptor: | L-fucose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Dang, S.Y, Wang, J, Yan, N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.143 Å) | | Cite: | Structure of a fucose transporter in an outward-open conformation
Nature, 467, 2010
|
|
5VO2
 
 | |
5VO1
 
 | |
6GSB
 
 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
2OUS
 
 | | crystal structure of PDE10A2 mutant D674A | | Descriptor: | MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUQ
 
 | | crystal structure of PDE10A2 in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7BTA
 
 | | Crystal structure of Rheb D60K mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTC
 
 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BU9
 
 | | Crystal Structure of Spindlin1-H3(K4me3-K9me2) complex | | Descriptor: | H3(K4me3-K9me2) peptide, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Molecular basis for histone H3 "K4me3-K9me3/2" methylation pattern readout by Spindlin1.
J.Biol.Chem., 295, 2020
|
|
7BQZ
 
 | |
7BTD
 
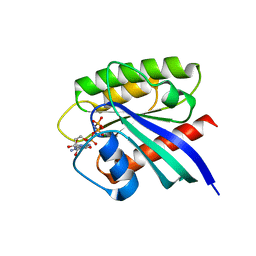 | | Crystal structure of Rheb Y35N mutant bound to GppNHp | | Descriptor: | GTP-binding protein Rheb, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
4C13
 
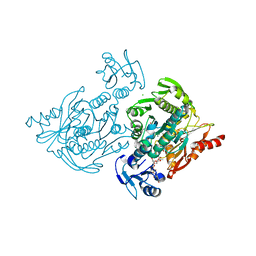 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
2L0K
 
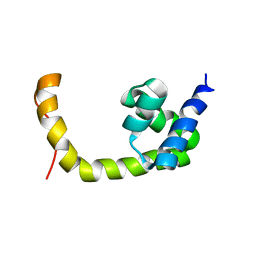 | | NMR solution structure of a transcription factor SpoIIID in complex with DNA | | Descriptor: | Stage III sporulation protein D | | Authors: | Chen, B, Himes, P, Lu, Z, Liu, A, Yan, H, Kroos, L. | | Deposit date: | 2010-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel Mode of DNA Binding by Bacterial Transcription Factor SpoIIID
To be Published
|
|
1C4G
 
 | |
5M93
 
 | |
5K4L
 
 | |
8JOP
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | Descriptor: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | Authors: | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | Deposit date: | 2023-06-08 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
