7R9G
 
 | | Catalytically inactive yeast Pseudouridine Synthase, PUS1, bound to RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*AP*AP*UP*CP*GP*GP*GP*AP*UP*UP*CP*CP*GP*GP*AP*UP*A)-3'), SULFATE ION, ... | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of mRNA recognition and binding by yeast pseudouridine synthase PUS1.
Plos One, 18, 2023
|
|
7R9F
 
 | |
2QOJ
 
 | | Coevolution of a homing endonuclease and its host target sequence | | Descriptor: | I-AniI DNA target seq1, I-AniI DNA target seq2, LAGLIDADG endonuclease, ... | | Authors: | Scalley-Kim, M, McConnell Smith, A, Stoddard, B.L. | | Deposit date: | 2007-07-20 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coevolution of a homing endonuclease and its host target sequence.
J.Mol.Biol., 372, 2007
|
|
2O3K
 
 | |
7RCF
 
 | |
7RCG
 
 | |
7RCD
 
 | |
7RCE
 
 | |
7RCC
 
 | |
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
3E0L
 
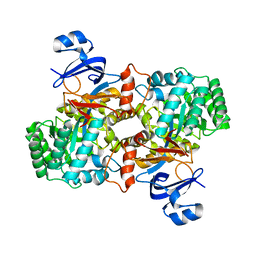 | | Computationally Designed Ammelide Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Murphy, P.M, Bolduc, J.M, Gallaher, J.L, Stoddard, B.L, Baker, D. | | Deposit date: | 2008-07-31 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Alteration of enzyme specificity by computational loop remodeling and design.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EH8
 
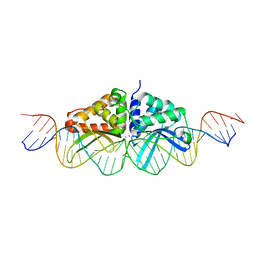 | |
3E54
 
 | | Archaeal Intron-encoded Homing Endonuclease I-Vdi141I Complexed With DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DGP*DAP*DCP*DTP*DCP*DTP*DCP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DGP*DGP*DCP*DTP*DAP*DCP*DCP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(P*DGP*DAP*DGP*DAP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Nomura, N, Nomura, Y, Sussman, D, Stoddard, B.L. | | Deposit date: | 2008-08-13 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of a common rDNA target site in archaea and eukarya by analogous LAGLIDADG and His-Cys box homing endonucleases
Nucleic Acids Res., 36, 2008
|
|
7SQ4
 
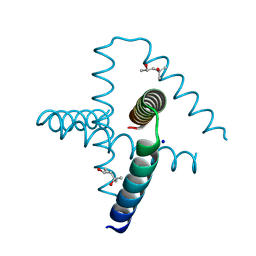 | | Designed trefoil knot protein, variant 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Designed trefoil knot protein, variant 2, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ3
 
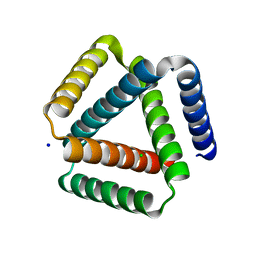 | | Designed trefoil knot protein, variant 1 | | Descriptor: | CHLORIDE ION, Designed trefoil knot protein, variant 1, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ5
 
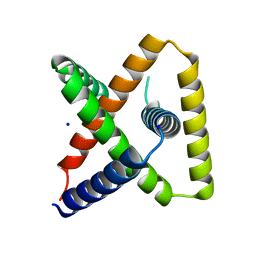 | | Designed trefoil knot protein, variant 3 | | Descriptor: | Designed trefoil knot protein, variant 3, SODIUM ION | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
4YIT
 
 | |
4YIS
 
 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4YHX
 
 | |
4Z20
 
 | |
4Z1Z
 
 | |
4YXX
 
 | |
6CZH
 
 | | Structure of a redesigned beta barrel, mFAP0, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP0 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
