4IOR
 
 | |
1ZOF
 
 | | Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter Pylori | | Descriptor: | alkyl hydroperoxide-reductase | | Authors: | Papinutto, E, Windle, H.J, Cendron, L, Battistutta, R, Kelleher, D, Zanotti, G. | | Deposit date: | 2005-05-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori.
Biochim.Biophys.Acta, 1753, 2005
|
|
4KWP
 
 | | Crystal Structure of Human CK2-alpha in complex with a benzimidazole inhibitor (K164) at 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4,5,6,7-tetrabromo-1-(2-deoxy-beta-D-erythro-pentofuranosyl)-1H-benzimidazole, Casein kinase II subunit alpha, ... | | Authors: | Ranchio, A, Lolli, G, Battistutta, R. | | Deposit date: | 2013-05-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Cell-permeable dual inhibitors of protein kinases CK2 and PIM-1: structural features and pharmacological potential.
Cell.Mol.Life Sci., 71, 2014
|
|
1S2X
 
 | | Crystal structure of Cag-Z from Helicobacter pylori | | Descriptor: | Cag-Z, ISOPROPYL ALCOHOL | | Authors: | Cendron, L, Seydel, A, Angelini, A, Battistutta, R, Zanotti, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of CagZ, a protein from the Helicobacter pylori pathogenicity island that encodes for a type IV secretion system
J.Mol.Biol., 340, 2004
|
|
6QS5
 
 | |
6RFE
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 4 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
6RFF
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 7 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-nitro-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
6RCM
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 3 | | Descriptor: | (~{E})-~{N}-(5-~{tert}-butyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
1NZE
 
 | | Crystal structure of PsbQ polypeptide of photosystem II from higher plants | | Descriptor: | Oxygen-evolving enhancer protein 3, ZINC ION | | Authors: | Calderone, V, Trabucco, M, Vujicic, A, Battistutta, R, Giacometti, G.M, Andreucci, F, Barbato, R, Zanotti, G. | | Deposit date: | 2003-02-17 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the PsbQ protein of photosystem II from higher plants
Embo Rep., 4, 2003
|
|
1OO2
 
 | | Crystal structure of transthyretin from Sparus aurata | | Descriptor: | CADMIUM ION, transthyretin | | Authors: | Pasquato, N, Ramazzina, I, Folli, C, Battistutta, R, Berni, R, Zanotti, G. | | Deposit date: | 2003-03-03 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Distinctive binding and structural properties of piscine transthyretin.
Febs Lett., 555, 2003
|
|
1M2P
 
 | | Crystal structure of 1,8-di-hydroxy-4-nitro-anthraquinone/CK2 kinase complex | | Descriptor: | 1,8-DI-HYDROXY-4-NITRO-ANTHRAQUINONE, Casein kinase II, alpha chain | | Authors: | De Moliner, E, Moro, S, Sarno, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
1M2R
 
 | | Crystal structure of 5,8-di-amino-1,4-di-hydroxy-anthraquinone/CK2 kinase complex | | Descriptor: | 5,8-DI-AMINO-1,4-DIHYDROXY-ANTHRAQUINONE, CASEIN kinase II, alpha chain | | Authors: | De Moliner, E, Moro, S, Sarno, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
1M2Q
 
 | | Crystal structure of 1,8-di-hydroxy-4-nitro-xanten-9-one/CK2 kinase complex | | Descriptor: | 1,8-DI-HYDROXY-4-NITRO-XANTHEN-9-ONE, Casein kinase II, alpha chain | | Authors: | De Moliner, E, Sarno, S, Moro, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
3PVG
 
 | | Crystal structure of Z. mays CK2 alpha subunit in complex with the inhibitor 4,5,6,7-tetrabromo-1-carboxymethylbenzimidazole (K68) | | Descriptor: | (4,5,6,7-tetrabromo-1H-benzimidazol-1-yl)acetic acid, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2010-12-07 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
3CWY
 
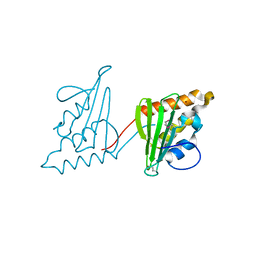 | | Structure of CagD from H. pylori pathogenicity island crystallized in the presence of Cu(II) ions | | Descriptor: | COPPER (II) ION, protein CagD | | Authors: | Cendron, L, Zanotti, G, Angelini, A, Barison, N, Couturier, M, Stein, M. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
J.Mol.Biol., 386, 2009
|
|
3CWX
 
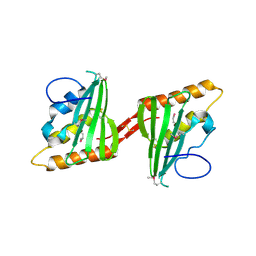 | | Crystal structure of cagd from helicobacter pylori pathogenicity island | | Descriptor: | protein CagD | | Authors: | Cendron, L, Zanotti, G, Angelini, A, Barison, N, Couturier, M, Stein, M. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
J.Mol.Biol., 386, 2009
|
|
1O9R
 
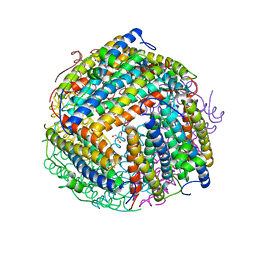 | | The X-ray crystal structure of Agrobacterium tumefaciens Dps, a member of the family that protect DNA without binding | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGROBACTERIUM TUMEFACIENS DPS, ... | | Authors: | Ilari, A, Ceci, P, Chiancone, E. | | Deposit date: | 2002-12-18 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Dps Protein of Agrobacterium Tumefaciens Does not Bind to DNA But Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties
J.Biol.Chem., 278, 2003
|
|
2PNC
 
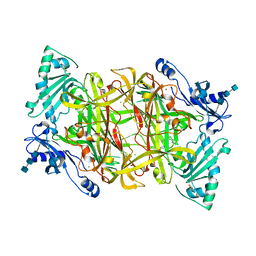 | | Crystal Structure of Bovine Plasma Copper-Containing Amine Oxidase in Complex with Clonidine | | Descriptor: | 2,6-DICHLORO-N-IMIDAZOLIDIN-2-YLIDENEANILINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Cendron, L, Holt, A, Smith, D.J, Zanotti, G, Rigo, A, Di Paolo, M.L. | | Deposit date: | 2007-04-24 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple binding sites for substrates and modulators of semicarbazide-sensitive amine oxidases: kinetic consequences
Mol.Pharmacol., 73, 2008
|
|
6RCB
 
 | |
6HOV
 
 | |
1LTR
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HUMAN HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SULFATE ION | | Authors: | Matkovic-Calogovic, D, Loreggian, A, Palu, G, Zanotti, G. | | Deposit date: | 1998-07-31 | | Release date: | 1999-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
2PYB
 
 | |
6ISJ
 
 | | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Cho, H.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans
To be published
|
|
1NA7
 
 | |
