7EMN
 
 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
7EPM
 
 | | human LDHC complexed with NAD+ and ethylamino acetic acid | | Descriptor: | 2-(ethylamino)-2-oxidanylidene-ethanoic acid, L-lactate dehydrogenase C chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-04-27 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of human LDHC4 as a potential target for anticancer drug discovery.
Acta Pharm Sin B, 12, 2022
|
|
8I8F
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed compound 1 | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-(2-naphthalen-1-yloxyethanoylamino)-2-oxidanyl-2-oxidanylidene-ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Shi, X, Liu, W. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8IEX
 
 | | Solution structure of AtWRKY11-DBD | | Descriptor: | Probable WRKY transcription factor 11, ZINC ION | | Authors: | Dong, X, Hu, Y.F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of Arabidopsis transcription factor WRKY11.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
7VK9
 
 | | Crystal structure of xCas9 P411T | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | Bao, R, Liu, H.Y, Luo, Y.Z, Song, Y.J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Acs Catalysis, 12, 2022
|
|
7FC3
 
 | | structure of NL63 receptor-binding domain complexed with horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X.Q, Ge, J.W, Lan, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7JU8
 
 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
7FC5
 
 | | Crystal structure of SARS-CoV-2 RBD and horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1 | | Authors: | Wang, X.Q, Lan, J, Ge, J.W. | | Deposit date: | 2021-07-13 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7LB7
 
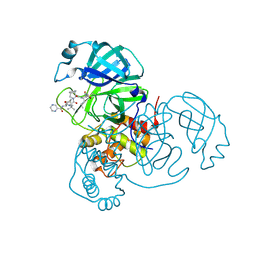 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Kneller, D.W, Coates, L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
J.Med.Chem., 64, 2021
|
|
8GRR
 
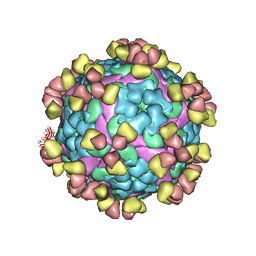 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W125 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Kun, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8GSP
 
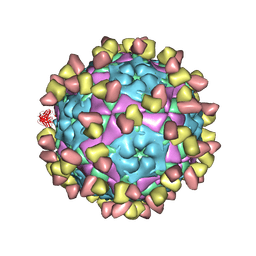 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W2 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8J19
 
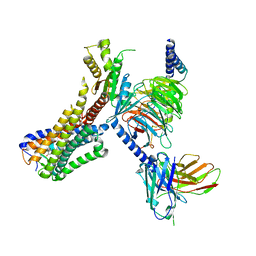 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J1A
 
 | | Cryo-EM structure of the GPR84 receptor-Gi complex with no ligand modeled | | Descriptor: | Antibody fragment ScFv16, G-protein coupled receptor 84, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
7FC6
 
 | | Crystal structure of SARS-CoV RBD and horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X.Q, Lan, J, Ge, J.W. | | Deposit date: | 2021-07-13 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7F29
 
 | |
4EVU
 
 | | Crystal structure of C-terminal domain of putative periplasmic protein ydgH from S. enterica | | Descriptor: | CHLORIDE ION, Putative periplasmic protein ydgH, SULFATE ION | | Authors: | Michalska, K, Cui, H, Xu, X, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Savchenko, A, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
4H1N
 
 | | Crystal Structure of P450 2B4 F297A Mutant in Complex with Anti-platelet Drug Clopidogrel | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Clopidogrel, Cytochrome P450 2B4, ... | | Authors: | Shah, M.B, Jang, H.H, Stout, C.D, Halpert, J.R. | | Deposit date: | 2012-09-10 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-ray crystal structure of the cytochrome P450 2B4 active site mutant F297A in complex with clopidogrel: Insights into compensatory rearrangements of the binding pocket.
Arch.Biochem.Biophys., 530, 2013
|
|
2JVA
 
 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | Descriptor: | Peptidyl-tRNA hydrolase domain protein | | Authors: | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XXI
 
 | | Crystal structure of CYP2C9 in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5YMU
 
 | |
5YSX
 
 | | Structure of P domain of GII.2 Noroviruses | | Descriptor: | VP1 | | Authors: | Duan, Z, Ao, Y. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Genetic Analysis of Reemerging GII.P16-GII.2 Noroviruses in 2016-2017 in China.
J. Infect. Dis., 218, 2018
|
|
2MJV
 
 | |
