2XDG
 
 | | Crystal structure of the extracellular domain of human growth hormone releasing hormone receptor. | | Descriptor: | 1,2-ETHANEDIOL, GROWTH HORMONE-RELEASING HORMONE RECEPTOR, MAGNESIUM ION | | Authors: | Pike, A.C.W, Quigley, A, Barr, A.J, Burgess Brown, N, Shrestha, L, Yang, J, Chaikuad, A, Vollmar, M, Muniz, J.R.C, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Carpenter, E.P. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Extracellular Domain of Human Growth Hormone Releasing Hormone Receptor.
To be Published
|
|
1OP9
 
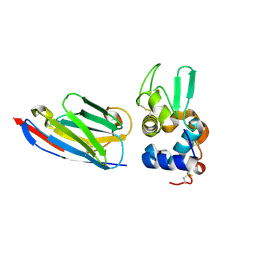 | | Complex of human lysozyme with camelid VHH HL6 antibody fragment | | Descriptor: | HL6 camel VHH fragment, Lysozyme C | | Authors: | Dumoulin, M, Last, A.M, Desmyter, A, Decanniere, K, Canet, D, Larsson, G, Spencer, A, Archer, D.B, Sasse, J, Muyldermans, S, Wyns, L, Redfield, C, Matagne, A, Robinson, C.V, Dobson, C.M. | | Deposit date: | 2003-03-05 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme
Nature, 424, 2003
|
|
3BIH
 
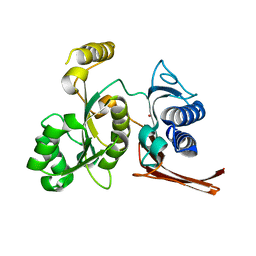 | | Crystal structure of fructose-1,6-bisphosphatase from E.coli GlpX | | Descriptor: | Fructose-1,6-bisphosphatase class II glpX, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A.F, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
2V30
 
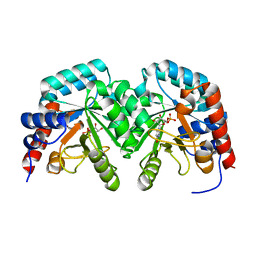 | | Human orotidine 5'-phosphate decarboxylase domain of uridine monophospate synthetase (UMPS) in complex with its product UMP. | | Descriptor: | OROTIDINE 5'-PHOSPHATE DECARBOXYLASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Moche, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Orotidine 5'-Decarboxylase Domain of Human Uridine Monophosphate Synthetase (Umps)
To be Published
|
|
4NWG
 
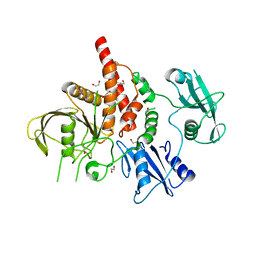 | | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation
To be Published
|
|
2AA1
 
 | | Crystal structure of the cathodic hemoglobin isolated from the Antarctic fish Trematomus Newnesi | | Descriptor: | Hemoglobin alpha-1 chain, Hemoglobin beta-C chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Bonomi, G, Lubrano, M.C, Merlino, A, Riccio, A, Vergara, A, Vitagliano, L, Verde, C, Di Prisco, G. | | Deposit date: | 2005-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Minimal structural requirements for root effect: crystal structure of the cathodic hemoglobin isolated from the antarctic fish Trematomus newnesi
Proteins, 62, 2006
|
|
4O1N
 
 | |
3S95
 
 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
1ZVJ
 
 | | Structure of Kumamolisin-AS mutant, D164N | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
2QES
 
 | |
2HTE
 
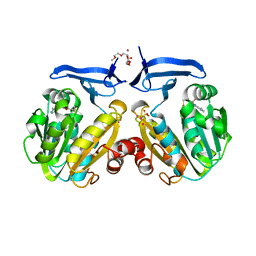 | | The crystal structure of spermidine synthase from p. falciparum in complex with 5'-methylthioadenosine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, SULFATE ION, ... | | Authors: | Qiu, W, Dong, A, Ren, H, Wu, H, Wasney, G, Vedadi, M, Lew, J, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Plotnikov, A.N, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
4O90
 
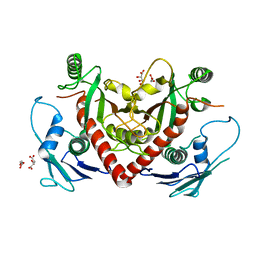 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.6A resolution | | Descriptor: | Chorismate synthase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Chaudhary, A, Singh, N, Shukla, P.K, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-31 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.6A resolution
To be Published
|
|
2OMA
 
 | |
4OHY
 
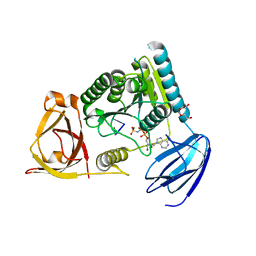 | | C. Elegans Clp1 bound to ssRNA dinucleotide GC, AMP-PNP, and Mg2+(inhibited substrate bound state) | | Descriptor: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
4OAV
 
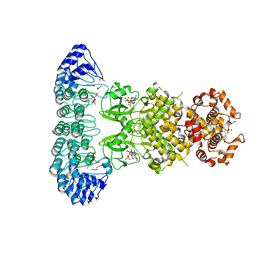 | | Complete human RNase L in complex with 2-5A (5'-ppp heptamer), AMPPCP and RNA substrate. | | Descriptor: | (2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-hydroxy-2-({[(S)-hydroxy{[(2R,3S,4S)-4-hydroxy-2-(hydroxymethyl)tetrahydrofuran-3-yl]oxy}phosphoryl]oxy}methyl)tetrahydrofuran-3-yl dihydrogen phosphate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
4OHV
 
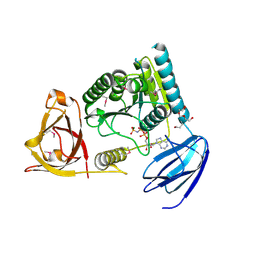 | | C. Elegans Clp1 bound to AMP-PNP, and Mg2+ | | Descriptor: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
4ONF
 
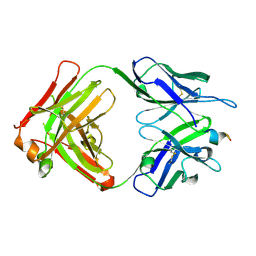 | | Fab fragment of 3D6 in complex with amyloid beta 1-7 | | Descriptor: | 3D6 FAB ANTIBODY HEAVY CHAIN, 3D6 FAB ANTIBODY LIGHT CHAIN, Amyloid beta A4 protein | | Authors: | Feinberg, H, Saldanha, J.W, Diep, L, Goel, A, Widom, A, Veldman, G.M, Weis, W.I, Schenk, D, Basi, G.S. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Alzheimers Res Ther, 6, 2014
|
|
2W9X
 
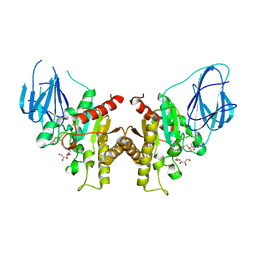 | | The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions | | Descriptor: | GLYCEROL, PUTATIVE ACETYL XYLAN ESTERASE | | Authors: | Montanier, C, Money, V.A, Pires, V, Flint, J.E, Benedita, P.A, Goyal, A, Prates, J.A, Izumi, A, Stalbrand, H, Morland, C, Cartmell, A, Kolenova, K, Topakas, E, Dobson, E, Bolam, D.N, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2W8G
 
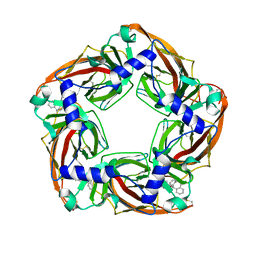 | | Aplysia californica AChBP bound to in silico compound 35 | | Descriptor: | (3-ENDO,8-ANTI)-8-BENZYL-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
4OY9
 
 | | Crystal structure of human P-Cadherin EC1-EC2 in closed conformation | | Descriptor: | CALCIUM ION, Cadherin-3 | | Authors: | Dalle Vedove, A, Lucarelli, A.P, Nardone, V, Matino, A, Parisini, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The X-ray structure of human P-cadherin EC1-EC2 in a closed conformation provides insight into the type I cadherin dimerization pathway.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4OZ7
 
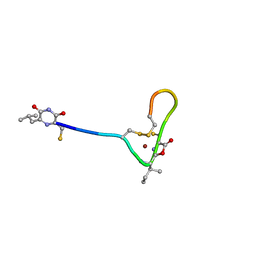 | |
4O7K
 
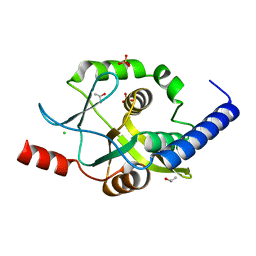 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, PHOSPHATE ION, ... | | Authors: | Maindola, P, Goyal, P, Arulandu, A. | | Deposit date: | 2013-12-25 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
2CPG
 
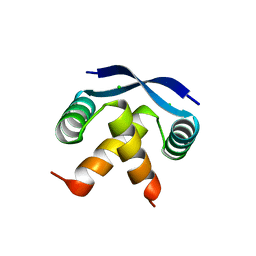 | | TRANSCRIPTIONAL REPRESSOR COPG | | Descriptor: | CHLORIDE ION, TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Sola, M, Acebo, P, Parraga, A, Guasch, A, Eritja, R, Gonzalez, A, Espinosa, M, del Solar, G, Coll, M. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator.
EMBO J., 17, 1998
|
|
3RAO
 
 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987. | | Descriptor: | Putative Luciferase-like Monooxygenase, SULFATE ION | | Authors: | Domagalski, M.J, Chruszcz, M, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987.
To be Published
|
|
