4IJU
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with (1S,4S)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol | | Descriptor: | (1s,4s)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol, CHLORIDE ION, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
3SQ1
 
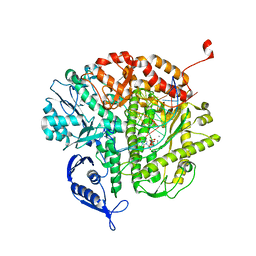 | | RB69 DNA Polymerase Ternary Complex with dUpCpp Opposite dA | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3', 5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | Descriptor: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
2WSO
 
 | | Structure of Cerulean Fluorescent Protein at physiological pH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Lelimousin, M, Noirclerc-Savoye, M, Lazareno-Saez, C, Paetzold, B, Le Vot, S, Chazal, R, Macheboeuf, P, Field, M.J, Bourgeois, D, Royant, A. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Intrinsic Dynamics in Ecfp and Cerulean Control Fluorescence Quantum Yield.
Biochemistry, 48, 2009
|
|
3SQ0
 
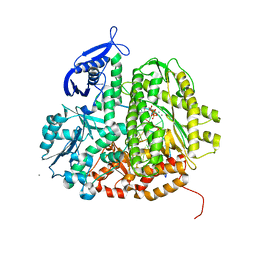 | | DNA Polymerase(L561A/S565G/Y567A) Ternary Complex with dUpNpp Opposite dA (Mn2+) | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
4IJV
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
7XMR
 
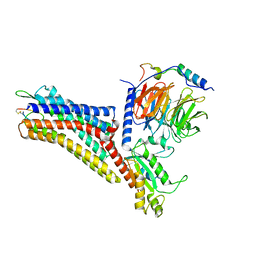 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
8HJE
 
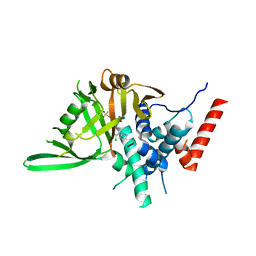 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
3SPZ
 
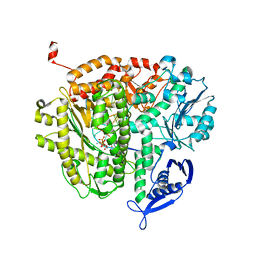 | | DNA Polymerase(L415A/L561A/S565G/Y567A) Ternary Complex with dUpCpp Opposite dA (Ca2+) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3', 5'-D(P*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
6IFU
 
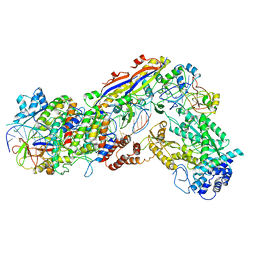 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | Descriptor: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
3BU7
 
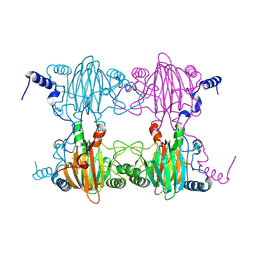 | | Crystal Structure and Biochemical Characterization of GDOsp, a Gentisate 1,2-Dioxygenase from Silicibacter Pomeroyi | | Descriptor: | FE (II) ION, Gentisate 1,2-dioxygenase | | Authors: | Chen, J, Wang, M.Z, Zhu, G.Y, Zhang, X.C, Rao, Z.H. | | Deposit date: | 2008-01-02 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and mutagenic analysis of GDOsp, a gentisate 1,2-dioxygenase from Silicibacter pomeroyi.
Protein Sci., 17, 2008
|
|
8J17
 
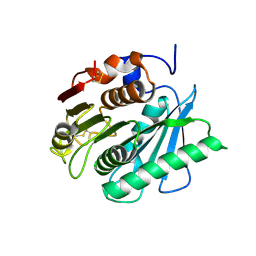 | | Crystal structure of IsPETase variant | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Yin, Q.D, Wang, Y.X. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Efficient polyethylene terephthalate biodegradation by an engineered Ideonella sakaiensis PETase with a fixed substrate-binding W156 residue
Green Chem, 2013
|
|
8WZ8
 
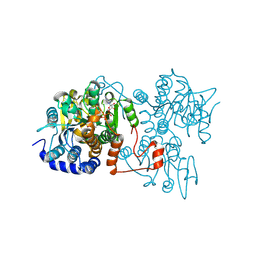 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD | | Descriptor: | Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WZ6
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and DZNep | | Descriptor: | Adenosylhomocysteinase, DZNep, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WZ7
 
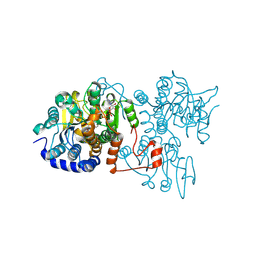 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(I255A,T287A) in ternary complex with NAD and adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8WWG
 
 | |
8WZ9
 
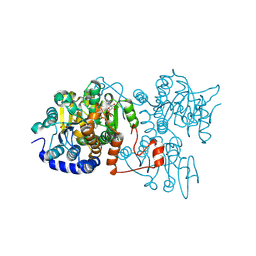 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and adenine | | Descriptor: | ADENINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis for substrate recognition by a S-adenosylhomocysteine hydrolase Lpg2021 from Legionella pneumophila.
Int.J.Biol.Macromol., 270, 2024
|
|
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 57, 2024
|
|
3QNN
 
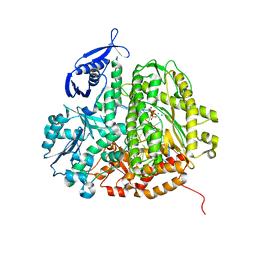 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dGT Opposite 3tCo | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA Primer, ... | | Authors: | Xia, S, Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Using a Fluorescent Cytosine Analogue tC(o) To Probe the Effect of the Y567 to Ala Substitution on the Preinsertion Steps of dNMP Incorporation by RB69 DNA Polymerase.
Biochemistry, 51, 2012
|
|
7YV4
 
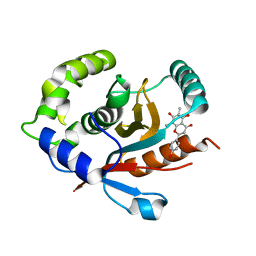 | | Crystal structure of human UCHL3 in complex with Farrerol | | Descriptor: | (2~{S})-2-(4-hydroxyphenyl)-6,8-dimethyl-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Mao, Z.Y, Xu, X.J, Zhang, W.T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Farrerol directly activates the deubiqutinase UCHL3 to promote DNA repair and reprogramming when mediated by somatic cell nuclear transfer.
Nat Commun, 14, 2023
|
|
8JED
 
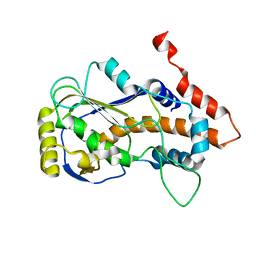 | | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors | | Descriptor: | mRNA-capping enzyme regulatory subunit OPG124 | | Authors: | Wang, D, Zhao, R, Shu, W, Hu, W, Wang, M, Cao, J, Zhou, X. | | Deposit date: | 2023-05-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors.
Int.J.Biol.Macromol., 253, 2023
|
|
