4E44
 
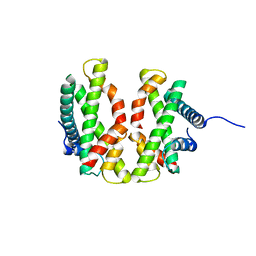 | |
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
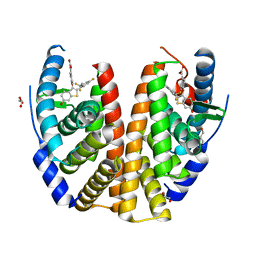
6B0F
 
 | |
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
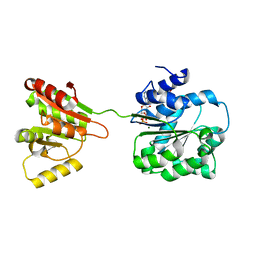 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
4DAY
 
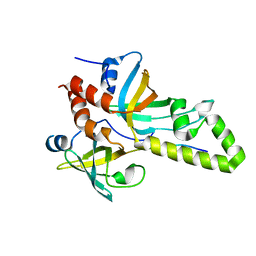 | | Crystal structure of the RMI core complex with MM2 peptide from FANCM | | Descriptor: | Fanconi anemia group M protein, RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Hoadley, K.A, Keck, J.L. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Defining the molecular interface that connects the Fanconi anemia protein FANCM to the Bloom syndrome dissolvasome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ARV
 
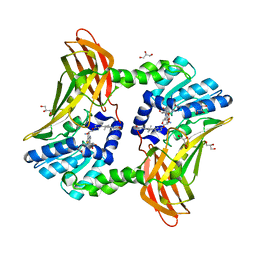 | | Crystal structure of CARM1 with Compound 2 and SAH | | Descriptor: | (2R)-1-amino-3-{3-[4-(morpholin-4-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-b]pyridin-6-yl]phenoxy}propan-2-ol, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.
Sci Rep, 7, 2017
|
|
5CES
 
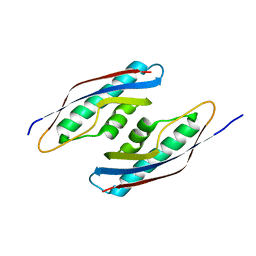 | |
6MCV
 
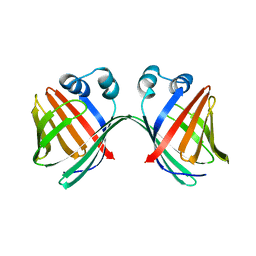 | |
4E45
 
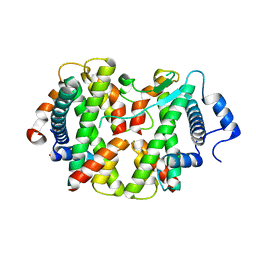 | | Crystal structure of the hMHF1/hMHF2 Histone-Fold Tetramer in Complex with Fanconi Anemia Associated Helicase hFANCM | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein, ... | | Authors: | Fox III, D, Zhao, Y, Yang, W, Weidong, W. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal that FANCM remodels the MHF Tetramer in favor of binding Branched DNA
To be Published
|
|
7WEU
 
 | |
7WET
 
 | | Crystal structure of Peroxiredoxin I in complex with the inhibitor Cela | | Descriptor: | (2R,4aS,6aS,12bR,14aS,14bR)-10-hydroxy-2,4a,6a,9,12b,14a-hexamethyl-11-oxo-1,2,3,4,4a,5,6,6a,11,12b,13,14,14a,14b-tetradecahydropicene-2-carboxylic acid, Peroxiredoxin-1 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2021-12-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1.
Signal Transduct Target Ther, 8, 2023
|
|
6SNJ
 
 | |
3JVS
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | 2-[(4-tert-butyl-3-nitrophenyl)carbonyl]-N-naphthalen-1-ylhydrazinecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
6BWG
 
 | |
5FQ1
 
 | |
3JVR
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
6MCU
 
 | |
6E51
 
 | |
6E50
 
 | |
6MKV
 
 | |
8RR2
 
 | | 3-keto-glycoside eliminase/hydratase in komplex with alpha-3-keto-glucose | | Descriptor: | (2~{R},3~{R},5~{S},6~{S})-2-(hydroxymethyl)-3,5,6-tris(oxidanyl)oxan-4-one, 1,2-ETHANEDIOL, 3-keto-disaccharide hydrolase domain-containing protein, ... | | Authors: | Pfeiffer, M, Kastner, K, Oberdorfer, G, Nidetzky, B. | | Deposit date: | 2024-01-22 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 2024
|
|
5V3B
 
 | |
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4KTE
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
