8I97
 
 | | Structure of Apo-C3aR-Go complex (Glacios) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I9A
 
 | | Structure of EP54-C3aR-Gq complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP54 ligand, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8I9S
 
 | | Structure of Apo-C3aR-Go complex (Titan) | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8IA2
 
 | | Structure of C5a bound human C5aR1 in complex with Go (Composite map) | | Descriptor: | Antibody fragment - ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8GOC
 
 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Vasopressin V2 receptor, V2R | | Descriptor: | Beta-arrestin-2, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GO8
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GP3
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GOO
 
 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-2, C5a anaphylatoxin chemotactic receptor 1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8I0N
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 (Local refine) | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8J6D
 
 | | Structure of EP141-C3aR-Go complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP141 peptide agonist, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-04-25 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
8J9K
 
 | | Structure of basal beta-arrestin2 | | Descriptor: | Beta-arrestin-2, Fab6 heavy chain, Fab6 light chain | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J8Z
 
 | | Structure of beta-arrestin1 in complex with D6Rpp | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J8V
 
 | | Structure of beta-arrestin2 in complex with D6Rpp (Local Refine) | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-2, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8JA3
 
 | | Structure of beta-arrestin1 in complex with C3aRpp | | Descriptor: | Beta-arrestin-1, C3a anaphylatoxin chemotactic receptor, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J97
 
 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local refine, cross-linked) | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8JAF
 
 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local Refine, non-cross linked) | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J8R
 
 | | Structure of beta-arrestin2 in complex with M2Rpp | | Descriptor: | Beta-arrestin-2, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8IY9
 
 | | Structure of Niacin-GPR109A-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYH
 
 | | Structure of MK6892-GPR109A-G-protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYW
 
 | | Structure of GSK256073-GPR109A-G-protein complex | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JHN
 
 | | Structure of MMF-GPR109A-G protein complex | | Descriptor: | (E)-4-methoxy-4-oxidanylidene-but-2-enoic acid, G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-24 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JER
 
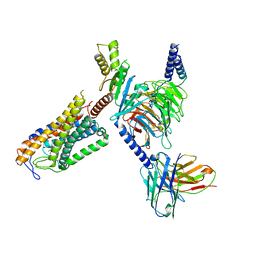 | | Structure of Acipimox-GPR109A-G protein complex | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JZZ
 
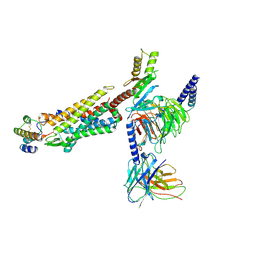 | | Structure of human C5a-desArg bound human C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
7U0P
 
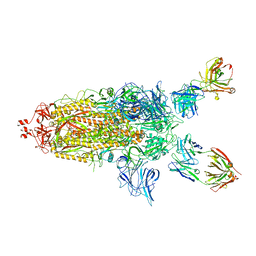 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7UPL
 
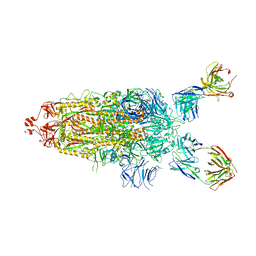 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
