7VGS
 
 | |
8XCD
 
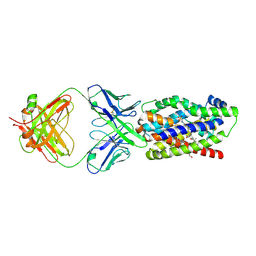 | | Macaca fascicularis NTCP in complex with YN69083 Fab | | Descriptor: | Solute carrier family 10 member a1, TAUROCHOLIC ACID, YN69083 Fab Heavy chain, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-12-08 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for hepatitis B virus restriction by a viral receptor homologue.
Nat Commun, 15, 2024
|
|
7W9W
 
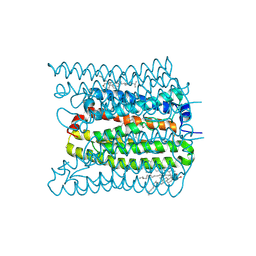 | | 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine | | Descriptor: | CHOLESTEROL, ChRmine, PALMITIC ACID, ... | | Authors: | Kishi, K.E, Kim, Y, Fukuda, M, Yamashita, K, Deisseroth, K, Kato, H.E. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine.
Cell, 185, 2022
|
|
6JOD
 
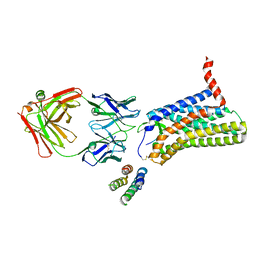 | | Angiotensin II type 2 receptor with ligand | | Descriptor: | Angiotensin II, Heavy chain of 4A03Fab, Light chain of 4A03Fab, ... | | Authors: | Asada, H, Iwata, S, Hirata, K, Shimamura, T. | | Deposit date: | 2019-03-20 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of Angiotensin II Type 2 Receptor with Endogenous Peptide Hormone.
Structure, 28, 2020
|
|
6K4J
 
 | | Crystal Structure of the the CD9 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | Authors: | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | Deposit date: | 2019-05-24 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
8J7X
 
 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7V
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7W
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J80
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 1, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-unbound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7U
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7T
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7Y
 
 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
7WU9
 
 | | Cryo-EM structure of the human EP3-Gi signaling complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suno, R, Sugita, Y, Morimoto, K, Iwasaki, K, Kato, T, Kobayashi, T. | | Deposit date: | 2022-02-07 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.375 Å) | | Cite: | Structural insights into the G protein selectivity revealed by the human EP3-G i signaling complex.
Cell Rep, 40, 2022
|
|
5YFI
 
 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YHL
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | Descriptor: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
6AAK
 
 | | Crystal structure of JAK3 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAH
 
 | | Crystal structure of JAK1 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
7D0I
 
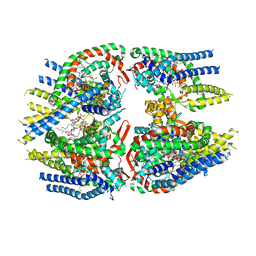 | | Cryo-EM structure of Schizosaccharomyces pombe Atg9 | | Descriptor: | Autophagy-related protein 9, Lauryl Maltose Neopentyl Glycol | | Authors: | Matoba, K, Tsutsumi, A, Kikkawa, M, Noda, N.N. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6AAJ
 
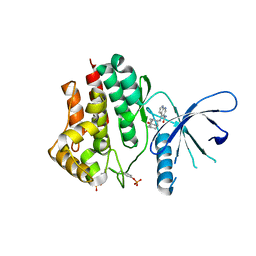 | | Crystal structure of JAK2 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
5YWY
 
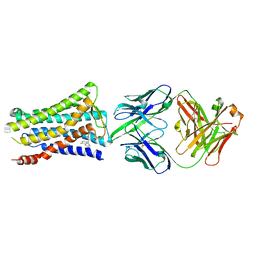 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | Descriptor: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
7DB6
 
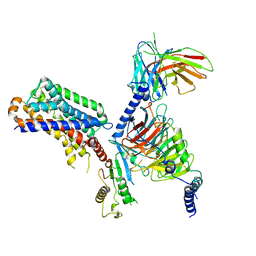 | | human melatonin receptor MT1 - Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Okamoto, H.H, Kusakizako, T, Shihioya, W, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human MT 1 -G i signaling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
