4O1O
 
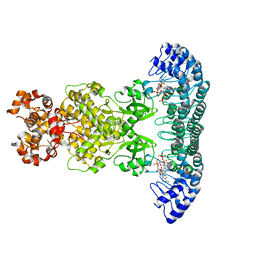 | | Crystal Structure of RNase L in complex with 2-5A | | Descriptor: | Ribonuclease L, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
3MT6
 
 | | Structure of ClpP from Escherichia coli in complex with ADEP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACYLDEPSIPEPTIDE 1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Chung, Y.S. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP.
Chem.Biol., 17, 2010
|
|
6CFD
 
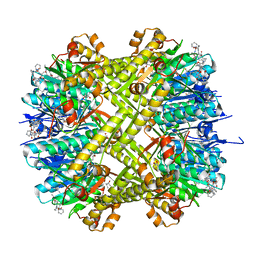 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
6NQB
 
 | |
5FBT
 
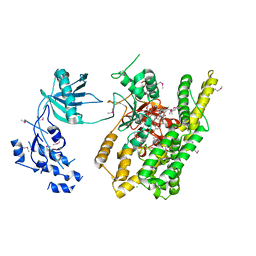 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin | | Descriptor: | CHLORIDE ION, Phosphoenolpyruvate synthase, Rifampin | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5FBS
 
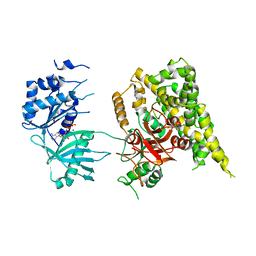 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with ADP and magnesium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoenolpyruvate synthase | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5VL3
 
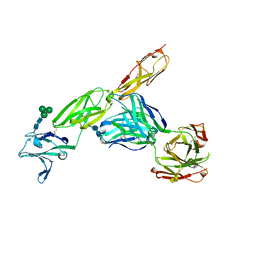 | | CD22 d1-d3 in complex with therapeutic Fab Epratuzumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell receptor CD22, Epratuzumab Fab Heavy Chain, ... | | Authors: | Sicard, T, Ereno-Orbea, J, Julien, J.P. | | Deposit date: | 2017-04-24 | | Release date: | 2017-10-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
5VKJ
 
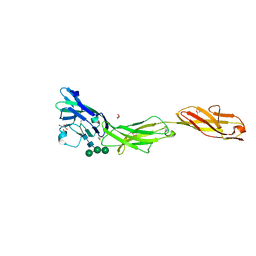 | | Crystal structure of human CD22 Ig domains 1-3 | | Descriptor: | B-cell receptor CD22, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Julien, J.P, Ereno-Orbea, J, Sicard, T. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
5VKK
 
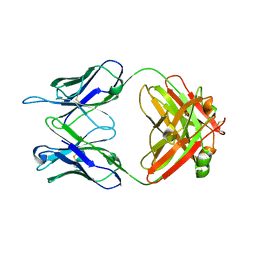 | |
5VKM
 
 | | Crystal structure of human CD22 Ig domains 1-3 in complex with alpha 2-6 sialyllactose | | Descriptor: | B-cell receptor CD22, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Julien, J.P, Ereno-Orbea, J, Sicard, T. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
6MG0
 
 | | Crystal structure of a 5-domain construct of LgrA in the thiolation state | | Descriptor: | 5'-({[(2R,3R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-{[oxido(oxo)phosphonio]oxy}butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-4-methylpentyl]sulfonyl}amino)-5'-deoxyadenosine, Linear gramicidin synthase subunit A | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFW
 
 | | Crystal structure of a 4-domain construct of LgrA in the substrate donation state | | Descriptor: | (2~{R})-~{N}-[3-[2-[[(2~{S})-2-formamido-3-methyl-butanoyl]amino]ethylamino]-3-oxidanylidene-propyl]-3,3-dimethyl-2-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-butanamide, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, ... | | Authors: | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFY
 
 | | Crystal structure of a 5-domain construct of LgrA in the substrate donation state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A, PHOSPHATE ION | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFX
 
 | | Crystal structure of a 4-domain construct of a mutant of LgrA in the substrate donation state | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFZ
 
 | | Crystal structure of dimodular LgrA in a condensation state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
5CW6
 
 | | Structure of metal dependent enzyme DrBRCC36 | | Descriptor: | DrBRCC36, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.193 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW4
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (Selenium Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, GLYCEROL, Protein FAM175B, ... | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW3
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (Zn Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW5
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (QSQ mutant) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.736 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
