3ZVE
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 84 | | Descriptor: | 3C PROTEASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVB
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 81 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-{[N-(TERT-BUTOXYCARBONYL)-L-PHENYLALANYL]AMINO}-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZV8
 
 | |
3ZVF
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 85 | | Descriptor: | 3C PROTEASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZV9
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 74 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVA
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 75 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVC
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 82 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(3-TERT-BUTOXY-3-OXOPROPYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVG
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | Descriptor: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVD
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 83 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
7P27
 
 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
3CDU
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
3CDW
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with protein primer VPg and a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
3ELY
 
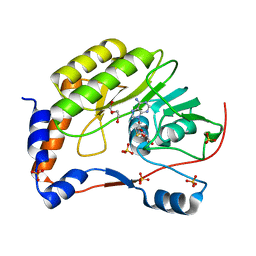 | | Wesselsbron virus Methyltransferase in complex with AdoHcy | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Bollati, M, Milani, M, Ricagno, S, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3EMB
 
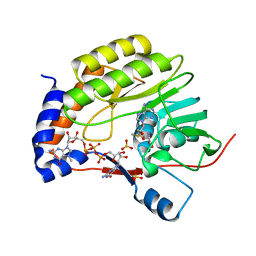 | | Wesselsbron virus Methyltransferase in complex with AdoMet and 7MeGpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Bollati, M, Ricagno, S, Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-24 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3ELW
 
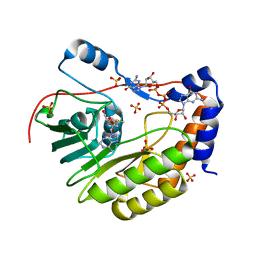 | | Wesselsbron virus Methyltransferase in complex with AdoMet and GpppG | | Descriptor: | DIGUANOSINE-5'-TRIPHOSPHATE, Methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Bollati, M, Ricagno, S, Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3ELU
 
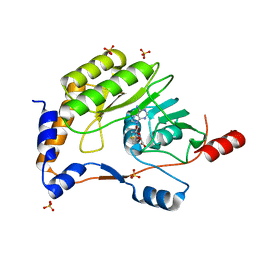 | | Wesselsbron virus Methyltransferase in complex with AdoMet | | Descriptor: | Methyltransferase, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Bollati, M, Ricagno, S, Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3EMD
 
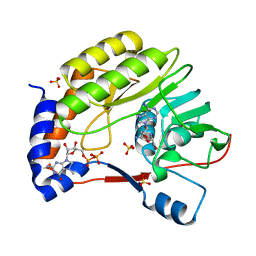 | | Wesselsbron virus Methyltransferase in complex with Sinefungin and 7MeGpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, SULFATE ION, ... | | Authors: | Bollati, M, Milani, M, Ricagno, S, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2008-09-24 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
3ELD
 
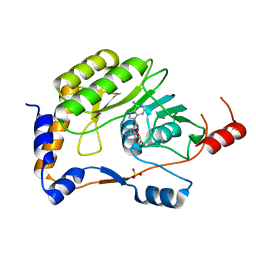 | | Wesselsbron methyltransferase in complex with Sinefungin | | Descriptor: | Methyltransferase, SINEFUNGIN, SULFATE ION | | Authors: | Bollati, M, Milani, M, Mastrangelo, E, Ricagno, S, Bolognesi, M. | | Deposit date: | 2008-09-22 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of RNA Cap in the Wesselsbron Virus NS5 Methyltransferase Domain: Implications for RNA-Capping Mechanisms in Flavivirus
J.Mol.Biol., 385, 2009
|
|
2WA1
 
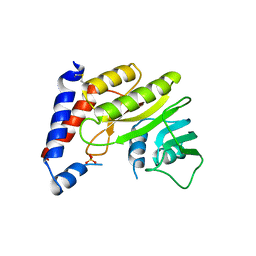 | | Structure of the methyltransferase domain from Modoc Virus, a Flavivirus with No Known Vector (NKV) | | Descriptor: | NON-STRUCTURAL PROTEIN 5, SULFATE ION | | Authors: | Jansson, A.M, Johansson, P, Jones, T.A. | | Deposit date: | 2009-02-02 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Methyltransferase Domain from the Modoc Virus, a Flavivirus with No Known Vector.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2WA2
 
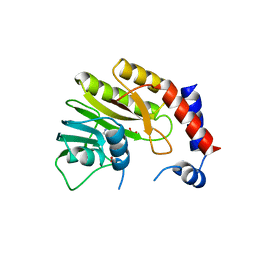 | | Structure of the methyltransferase domain from Modoc Virus, a Flavivirus with No Known Vector (NKV) | | Descriptor: | NON-STRUCTURAL PROTEIN 5, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Jansson, A.M, Johansson, P, Jones, T.A. | | Deposit date: | 2009-02-02 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Methyltransferase Domain from the Modoc Virus, a Flavivirus with No Known Vector.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5E9Q
 
 | |
5EIF
 
 | |
5EHG
 
 | |
5EKX
 
 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYLMETHIONINE AND FRAGMENT NB2E11 | | Descriptor: | 4-chloro-5-methylbenzene-1,2-diamine, NS5 METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Barral, K, Bricogne, G, Sharff, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design.
Eur.J.Med.Chem., 125, 2016
|
|
5EHI
 
 | |
