3TU4
 
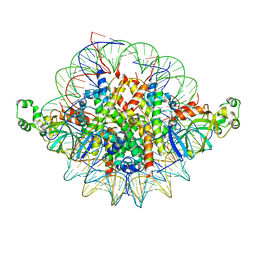 | | Crystal structure of the Sir3 BAH domain in complex with a nucleosome core particle. | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Armache, K.-J, Garlick, J.D, Canzio, D, Narlikar, G.J, Kingston, R.E. | | Deposit date: | 2011-09-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution.
Science, 334, 2011
|
|
8C86
 
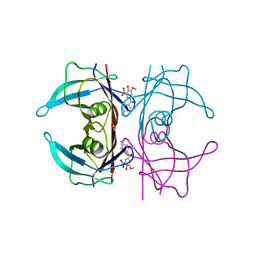 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone analogue 2 | | Descriptor: | (2,4-dimethylphenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Poonsiri, T, Benini, S, Loconte, V, Cianci, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 3-O-Methyltolcapone and Its Lipophilic Analogues Are Potent Inhibitors of Transthyretin Amyloidogenesis with High Permeability and Low Toxicity.
Int J Mol Sci, 25, 2023
|
|
8C85
 
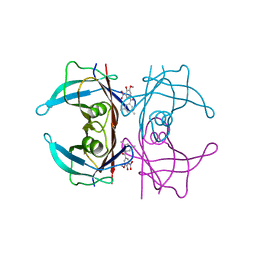 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone analogue 1 | | Descriptor: | (3,5-dimethylphenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Poonsiri, T, Benini, S, Loconte, V, Cianci, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | 3-O-Methyltolcapone and Its Lipophilic Analogues Are Potent Inhibitors of Transthyretin Amyloidogenesis with High Permeability and Low Toxicity.
Int J Mol Sci, 25, 2023
|
|
5NSQ
 
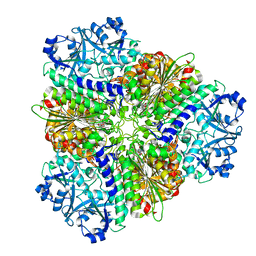 | |
5NTG
 
 | |
5NSK
 
 | | apo Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative | | Authors: | Timm, J, Wilson, K.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
5NTF
 
 | | apo Structure of Leucyl aminopeptidase from Trypanosoma cruzi | | Descriptor: | Aminopeptidase, SULFATE ION | | Authors: | Timm, J, Wilson, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2017
|
|
5NTH
 
 | |
5NTD
 
 | | Structure of Leucyl aminopeptidase from Trypanosoma brucei in complex with Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BICARBONATE ION, ... | | Authors: | Timm, J, Wilson, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
5NSM
 
 | | unliganded Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative, BICARBONATE ION, ... | | Authors: | Timm, J, Wilson, K. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
5C9C
 
 | | CRYSTAL STRUCTURE OF BRAF(V600E) IN COMPLEX WITH LY3009120 COMPND | | Descriptor: | 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Edwards, T, Abendroth, J, Chun, L. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers.
Cancer Cell, 28, 2015
|
|
7L3N
 
 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
7KMG
 
 | | LY-CoV555 neutralizing antibody against SARS-CoV-2 | | Descriptor: | GLYCEROL, LY-CoV555 Fab heavy chain, LY-CoV555 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMH
 
 | | LY-CoV488 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV488 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Boyles, J.S, Dickinson, C.D, Coleman, K.A. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMI
 
 | | LY-CoV481 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV481 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
4QQC
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Domain in Complex with FIIN-2, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR Kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R6V
 
 | | Crystal Structure of FGF Receptor (FGFR) 4 Kinase Harboring the V550L Gate-Keeper Mutation in Complex with FIIN-3, an Irreversible Tyrosine Kinase Inhibitor Capable of Overcoming FGFR kinase Gate-Keeper Mutations | | Descriptor: | Fibroblast growth factor receptor 4, N-[4-({[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino}methyl)phenyl]propanamide, SULFATE ION | | Authors: | Huang, Z, Mohammadi, M. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
7Q4A
 
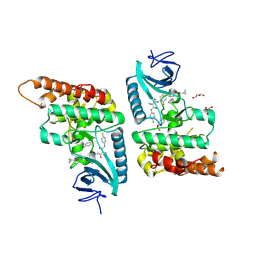 | | Toxoplasma gondii PRP4K kinase domain (L715F) bound to altiratinib | | Descriptor: | Altiratinib, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Swale, C, Bellini, V, Bowler, M. | | Deposit date: | 2021-10-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Altiratinib blocks Toxoplasma gondii and Plasmodium falciparum development by selectively targeting a spliceosome kinase.
Sci Transl Med, 14, 2022
|
|
3IKA
 
 | |
6DO3
 
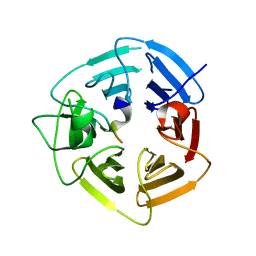 | | KLHDC2 ubiquitin ligase in complex with SelK C-end degron | | Descriptor: | Kelch domain-containing protein 2, SelK C-end Degron | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
8UXS
 
 | |
5D41
 
 | | EGFR kinase domain in complex with mutant selective allosteric inhibitor | | Descriptor: | (2R)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-2-phenyl-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Yun, C.-H, Park, E, Eck, M.J. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors.
Nature, 534, 2016
|
|
6DO5
 
 | | KLHDC2 ubiquitin ligase in complex with USP1 C-end degron | | Descriptor: | Kelch domain-containing protein 2, USP1 C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
6DO4
 
 | | KLHDC2 ubiquitin ligase in complex with SelS C-end degron | | Descriptor: | Kelch domain-containing protein 2, SELS C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
