7Y59
 
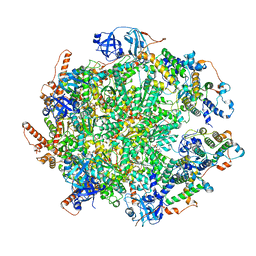 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
5VRN
 
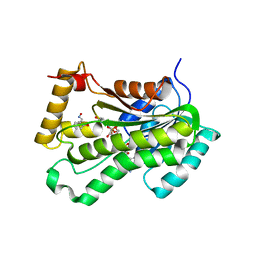 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-4-[[5-[4-cyano-2-[(~{E})-hydroxyiminomethyl]phenoxy]-1-oxidanyl-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5YGI
 
 | |
5VRL
 
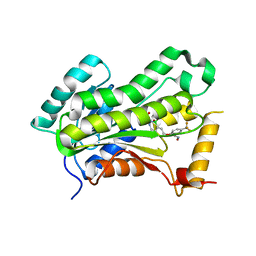 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | (~{N}~{E})-~{N}-[[2-[[2-ethylsulfonyl-1,1-bis(oxidanyl)-3,4-dihydro-2,3,1$l^{4}-benzodiazaborinin-7-yl]oxy]-5-(trifluoromethyl)phenyl]methylidene]hydroxylamine, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5VRM
 
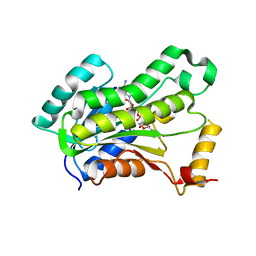 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3-oxidanyl-4-[[1-oxidanyl-6-[4-(trifluoromethyl)phenoxy]-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
8OUW
 
 | | Cryo-EM structure of CMG helicase bound to TIM-1/TIPN-1 and homodimeric DNSN-1 on fork DNA (Caenorhabditis elegans) | | Descriptor: | Cell division control protein 45 homolog, DNA Lagging Strand Template, DNA Leading Strand Template, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Labib, K.P.M. | | Deposit date: | 2023-04-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | DNSN-1 recruits GINS for CMG helicase assembly during DNA replication initiation in Caenorhabditis elegans.
Science, 381, 2023
|
|
7YUD
 
 | | FTY720p-bound human SPNS2 | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, NbFab L-chain, NbFab-H-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUB
 
 | | S1P-bound human SPNS2 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, NbFab-H-chain, NbFab-L-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUF
 
 | | apo human SPNS2 | | Descriptor: | NbFab H-chain, NbFab L-chain, Sphingosine-1-phosphate transporter SPNS2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
7Q2U
 
 | | The crystal structure of the HINT1 Q62A mutant. | | Descriptor: | CACODYLATE ION, HEXAETHYLENE GLYCOL, Histidine triad nucleotide-binding protein 1, ... | | Authors: | Dolot, R.M, Strom, A.M, Wagner, C.R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dynamic Long-Range Interactions Influence Substrate Binding and Catalysis by Human Histidine Triad Nucleotide-Binding Proteins (HINTs), Key Regulators of Multiple Cellular Processes and Activators of Antiviral ProTides.
Biochemistry, 61, 2022
|
|
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
8W9Y
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | Descriptor: | Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
8HZW
 
 | | The NMR structure of noursinH11W peptide | | Descriptor: | noursinH11W | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
4U93
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4W7T
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
8KAE
 
 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
4ZS3
 
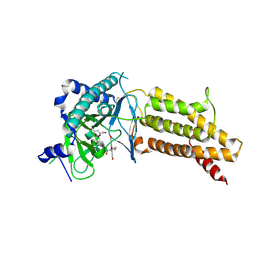 | | Structural complex of 5-aminofluorescein bound to the FTO protein | | Descriptor: | 2-OXOGLUTARIC ACID, 5-amino-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-11-04 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
4ZS2
 
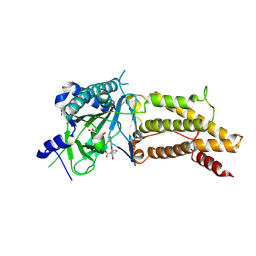 | | Structural complex of FTO/fluorescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-11-04 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
3K30
 
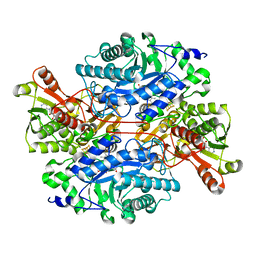 | | Histamine dehydrogenase from Nocardiodes simplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Histamine dehydrogenase, ... | | Authors: | Scott, E.E, Reed, T.M, Limburg, J. | | Deposit date: | 2009-09-30 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histamine dehydrogenase from Nocardioides simplex.
J.Biol.Chem., 285, 2010
|
|
4KM9
 
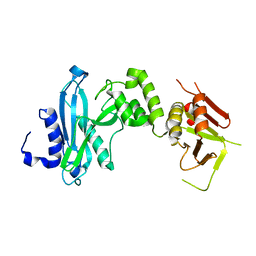 | |
4KM8
 
 | | Crystal structure of Sufud60 | | Descriptor: | Suppressor of fused homolog | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
4KMH
 
 | |
4KMD
 
 | | Crystal structure of Sufud60-Gli1p | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Sufu, ... | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
