4MSG
 
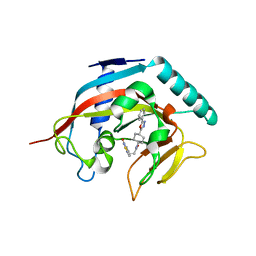 | | Crystal structure of tankyrase 1 with compound 22 | | Descriptor: | 3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]-N-[trans-4-(5-phenyl-1,3,4-oxadiazol-2-yl)cyclohexyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4P7S
 
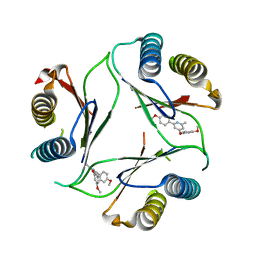 | |
7NDX
 
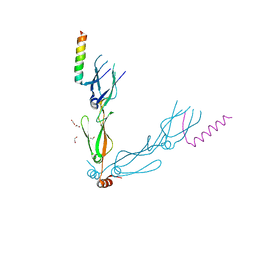 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
4OGP
 
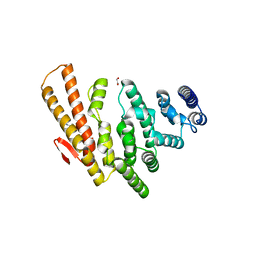 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4MT9
 
 | | Co-crystal structure of tankyrase 1 with compound 49 | | Descriptor: | N-[trans-4-(4-cyanophenoxy)cyclohexyl]-3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-19 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4N4V
 
 | |
4PKZ
 
 | |
7MN7
 
 | | PTP1B F225Y in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MN9
 
 | | PTP1B 1-284 F225Y-R199N | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNA
 
 | | PTP1B 1-284 F225Y-R199N in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNC
 
 | | PTP1B L204A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MND
 
 | | PTP1B L204A in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNF
 
 | | PTP1B P206G in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNE
 
 | | PTP1B P206G mutation, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MNB
 
 | | PTP1B F225Y-R199N-L195R in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MOW
 
 | | PTP1B F225I in complex with TCS401 | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MOV
 
 | | PTP1B 1-301 F225Y-R199N mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MOU
 
 | | PTP1B F225Y-R199N-L195R | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
7MKZ
 
 | | PTP1B F225Y mutant, open state | | Descriptor: | CHLORIDE ION, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Torgeson, K.R, Page, R, Peti, W. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conserved conformational dynamics determine enzyme activity.
Sci Adv, 8, 2022
|
|
4PLU
 
 | |
4MSK
 
 | | Co-crystal structure of tankyrase 1 with compound 34 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[4-(5-phenyl-1,3,4-oxadiazol-2-yl)phenyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4N4T
 
 | |
6DAU
 
 | | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae | | Descriptor: | GLYCEROL, Spermidine N1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Beahan, A, Kulyavtsev, P, Tan, L, Tran, D, Kuhn, M.L, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To be Published
|
|
1BMB
 
 | | GRB2-SH2 DOMAIN IN COMPLEX WITH KPFY*VNVEF (PKF270-974) | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR BOUND PROTEIN 2), PROTEIN (PKF270-974) | | Authors: | Rondeau, J.M, Zurini, M. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1).
J.Med.Chem., 42, 1999
|
|
1BM2
 
 | |
