3U83
 
 | | Crystal structure of nectin-1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
3U82
 
 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | Descriptor: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.164 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
1Q2K
 
 | | Solution structure of BmBKTx1 a new potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Neurotoxin BmK37 | | Authors: | Cai, Z, Xu, C, Xu, Y, Lu, W, Chi, C.W, Shi, Y, Wu, J. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmBKTx1, a New BK(Ca)(1) Channel Blocker from the Chinese Scorpion Buthus martensi Karsch(,).
Biochemistry, 43, 2004
|
|
1K99
 
 | |
4M69
 
 | | Crystal structure of the mouse RIP3-MLKL complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mixed lineage kinase domain-like protein, ... | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4M68
 
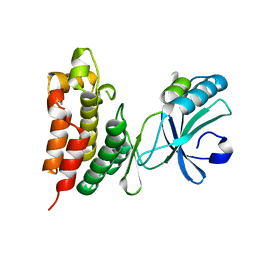 | | Crystal structure of the mouse MLKL kinase-like domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
1L8Z
 
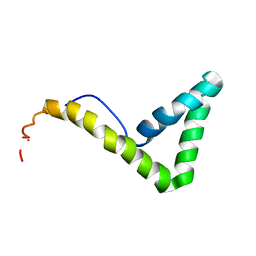 | | Solution structure of HMG box 5 in human upstream binding factor | | Descriptor: | upstream binding factor 1 | | Authors: | Yang, W, Xu, Y, Wu, J, Zeng, W, Shi, Y. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor
Biochemistry, 42, 2003
|
|
4LLA
 
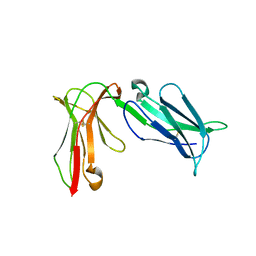 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
4LNR
 
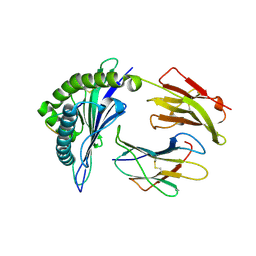 | | The structure of HLA-B*35:01 in complex with the peptide (RPQVPLRPMTY) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Cheng, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-12 | | Release date: | 2014-07-23 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex.
J.Biol.Chem., 289, 2014
|
|
4LL9
 
 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
1L8Y
 
 | | Solution structure of HMG box 5 in human upstream binding factor | | Descriptor: | upstream binding factor 1 | | Authors: | Yang, W, Xu, Y, Wu, J, Zeng, W, Shi, Y. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor
Biochemistry, 42, 2003
|
|
4M66
 
 | | Crystal structure of the mouse RIP3 kinase domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
1OZJ
 
 | | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | | Descriptor: | SMAD 3, Smad binding element, ZINC ION | | Authors: | Chai, J, Wu, J.-W, Yan, N, Massague, J, Pavletich, N.P, Shi, Y. | | Deposit date: | 2003-04-09 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Features of a Smad3 MH1-DNA complex. Roles of water and zinc in DNA binding.
J.Biol.Chem., 278, 2003
|
|
4KMP
 
 | | Structure of XIAP-BIR3 and inhibitor | | Descriptor: | (2S,2'S)-N,N'-[(6,6'-difluoro-1H,1'H-2,2'-biindole-3,3'-diyl)bis{methanediyl[(2R,4S)-4-hydroxypyrrolidine-2,1-diyl][(2S)-1-oxobutane-1,2-diyl]}]bis[2-(methylamino)propanamide], E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Li, X, Wang, J, Condon, S.M, Shi, Y. | | Deposit date: | 2013-05-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of XIAP-BIR3 and inhibitor
To be Published
|
|
4M67
 
 | | Crystal structure of the human MLKL kinase-like domain | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
1RJI
 
 | | Solution Structure of BmKX, a novel potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | potassium channel toxin KX | | Authors: | Cai, Z, Wu, J, Xu, Y, Wang, C.-G, Chi, C.-W, Shi, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A novel short-chain peptide BmKX from the Chinese scorpion Buthus martensi karsch, sequencing, gene cloning and structure determination
Toxicon, 45, 2005
|
|
4KMN
 
 | | Structure of cIAP1-BIR3 and inhibitor | | Descriptor: | (2S)-N-{(2R)-1-[(2R,4S)-2-{[6,6'-difluoro-3'-({(2R,4S)-4-hydroxy-1-[(2S)-2-{[(2S)-2-(methylamino)propanoyl]amino}butanoyl]pyrrolidin-2-yl}methyl)-1H,1'H-2,2'-biindol-3-yl]methyl}-4-hydroxypyrrolidin-1-yl]-1-oxobutan-2-yl}-2-(methylamino)propanamide, Baculoviral IAP repeat-containing protein 2, PHOSPHATE ION, ... | | Authors: | Li, X, Wang, J, Condon, S.M, Shi, Y. | | Deposit date: | 2013-05-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure of cIAP1-BIR3 and inhibitor
To be Published
|
|
4O2C
 
 | | An Nt-acetylated peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
4O2F
 
 | | A peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
4O2E
 
 | | A peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
4O79
 
 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 10 minutes | | Descriptor: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7W5A
 
 | | The cryo-EM structure of human pre-C*-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W5B
 
 | | The cryo-EM structure of human C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W59
 
 | | The cryo-EM structure of human pre-C*-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
8X54
 
 | | Cryo-EM structure of human gamma-secretase in complex with APP-C99 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
