4JY2
 
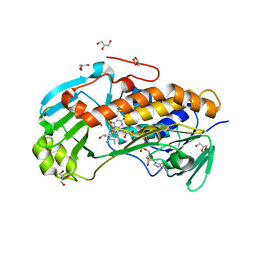 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, native and unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
7T3P
 
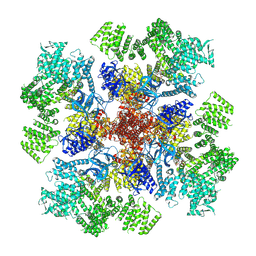 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active A state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3Q
 
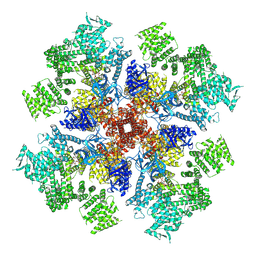 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active B state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3T
 
 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3U
 
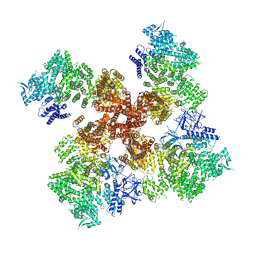 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3R
 
 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active C state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
5IX7
 
 | | Crystal structure of metallo-DNA nanowire with infinite one-dimensional silver array | | Descriptor: | DNA (5'-D(*GP*GP*AP*CP*TP*(CBR)P*GP*AP*CP*TP*CP*C)-3'), POTASSIUM ION, SILVER ION | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Hattori, Y, Saneyoshi, H, Ono, A, Tanaka, Y. | | Deposit date: | 2016-03-23 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A metallo-DNA nanowire with uninterrupted one-dimensional silver array
Nat Chem, 9, 2017
|
|
3B1U
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-F-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-fluoroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
4JY3
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-pyridoxic acid bound form | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-4-(hydroxymethyl)-6-methylpyridine-3-carboxylic acid, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-29 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
3B1T
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-Cl-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-chloroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
5KVB
 
 | |
1MG8
 
 | | NMR structure of ubiquitin-like domain in murine Parkin | | Descriptor: | Parkin | | Authors: | Tashiro, M, Okubo, S, Shimotakahara, S, Hatanaka, H, Yasuda, H, Kainosho, M, Yokoyama, S, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of ubiquitin-like domain in PARKIN: Gene product of familial Parkinson's disease.
J.Biomol.NMR, 25, 2003
|
|
1DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
5B2T
 
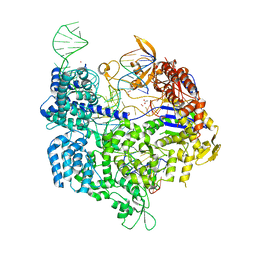 | | Crystal structure of the Streptococcus pyogenes Cas9 VRER variant in complex with sgRNA and target DNA (TGCG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2S
 
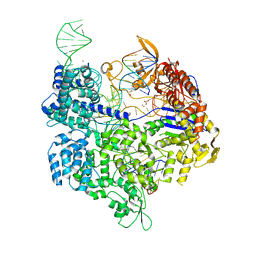 | | Crystal structure of the Streptococcus pyogenes Cas9 EQR variant in complex with sgRNA and target DNA (TGAG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B4C
 
 | | Crystal structure of H10N mutant of LpxH with manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
6LKE
 
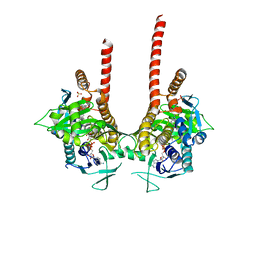 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
5EGE
 
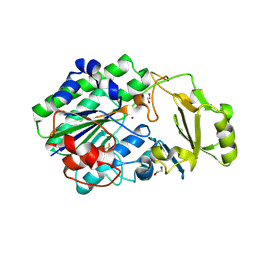 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
5B4B
 
 | | Crystal structure of LpxH with lipid X in spacegroup C2 | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
5B49
 
 | | Crystal structure of LpxH with manganese from Pseudomonas aeruginosa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
5B4D
 
 | | Crystal structure of H10N mutant of LpxH | | Descriptor: | GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
6MR4
 
 | | Crystal structure of the Sth1 bromodomain from S.cerevisiae | | Descriptor: | Nuclear protein STH1/NPS1 | | Authors: | Seo, H.S, Hashimoto, H, Krolak, A, Debler, E.W, Blus, B.J. | | Deposit date: | 2018-10-11 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Substrate Affinity and Specificity of the ScSth1p Bromodomain Are Fine-Tuned for Versatile Histone Recognition.
Structure, 27, 2019
|
|
5EGH
 
 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase in complex with phosphocholine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
1KWG
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trimeric Crystal Structure of the Glycoside Hydrolase Family 42 beta-Galactosidase from Thermus thermophilus A4 and the Structure of its Complex with Galactose
J.MOL.BIOL., 322, 2002
|
|
