4IVD
 
 | | JAK1 kinase (JH1 domain) in complex with compound 34 | | Descriptor: | 3-(trans-4-{2-[(1R)-1-hydroxyethyl]imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl}cyclohexyl)propanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2013-01-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identification of C-2 Hydroxyethyl Imidazopyrrolopyridines as Potent JAK1 Inhibitors with Favorable Physicochemical Properties and High Selectivity over JAK2.
J.Med.Chem., 56, 2013
|
|
4IVA
 
 | | JAK2 kinase (JH1 domain) in complex with the inhibitor TRANS-4-[(8AS)-2-[(1R)-1-HYDROXYETHYL]IMIDAZO[4,5-D]PYRROLO[2,3-B]PYRIDIN-1(8AH)-YL]CYCLOHEXANECARBONITRILE | | Descriptor: | Tyrosine-protein kinase JAK2, trans-4-{2-[(1R)-1-hydroxyethyl]imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl}cyclohexanecarbonitrile | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-01-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of C-2 Hydroxyethyl Imidazopyrrolopyridines as Potent JAK1 Inhibitors with Favorable Physicochemical Properties and High Selectivity over JAK2.
J.Med.Chem., 56, 2013
|
|
8FTI
 
 | |
3VWB
 
 | | Crystal structure of VirB core domain (Se-Met derivative) complexed with the cis-acting site (5-BRU modifications) upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator virB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
3W2A
 
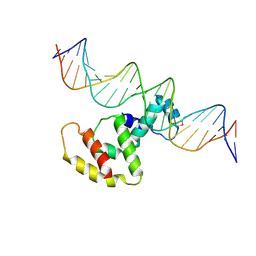 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsp promoter | | Descriptor: | DNA (31-mer), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-11-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.775 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
3W3C
 
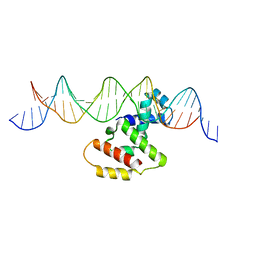 | | Crystal structure of VirB core domain complexed with the cis-acting site upstream icsb promoter | | Descriptor: | DNA (26-MER), Virulence regulon transcriptional activator VirB | | Authors: | Gao, X.P, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes
Nucleic Acids Res., 41, 2013
|
|
5BMV
 
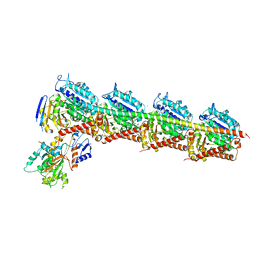 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-Vinblastine COMPLEX | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Chen, Q, Zhang, R. | | Deposit date: | 2015-05-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules.
Mol.Pharmacol., 89, 2016
|
|
6NNG
 
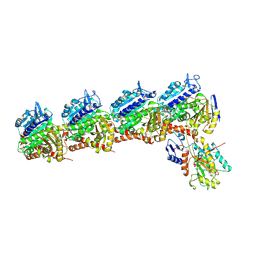 | | Tubulin-RB3_SLD-TTL in complex with compound DJ95 | | Descriptor: | 2-(1H-indol-6-yl)-4-(3,4,5-trimethoxyphenyl)-1H-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Colchicine Binding Site Agent DJ95 Overcomes Drug Resistance and Exhibits Antitumor Efficacy.
Mol.Pharmacol., 96, 2019
|
|
6O3N
 
 | |
7Y7J
 
 | | SARS-CoV-2 S trimer in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
 | | SARS-CoV-2 RBD in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, Spike protein S1 | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
4FHA
 
 | | Structure of Dihydrodipicolinate Synthase from Streptococcus pneumoniae,bound to pyruvate and lysine | | Descriptor: | Dihydrodipicolinate synthase, LYSINE, SODIUM ION | | Authors: | Perugini, M.A, Dogovski, C, Parker, M.W, Gorman, M.A. | | Deposit date: | 2012-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure, Function, Stability and Knockout Phenotype of Dihydrodipicolinate Synthase from Streptococcus pneumoniae
To be Published
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7KPT
 
 | | Crystal structure of CtdE in complex with FAD and substrate 4 | | Descriptor: | (6aR,7aS,11S,13aS)-6,6,11-trimethyl-4-(3-methylbut-2-en-1-yl)-6,6a,7,8,9,10,11,14-octahydro-5H,13H-13a,7a-(epiminomethano)quinolizino[2,3-b]carbazol-16-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
7KPQ
 
 | | Crystal structure of CtdE in complex with FAD | | Descriptor: | FAD-dependent monooxygenase CtdE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhao, B, Hu, L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins.
Nat Commun, 12, 2021
|
|
8GZJ
 
 | |
8GZL
 
 | |
8GZM
 
 | |
8GZK
 
 | |
3E5A
 
 | | Crystal structure of Aurora A in complex with VX-680 and TPX2 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, SULFATE ION, Serine/threonine-protein kinase 6, ... | | Authors: | Zhao, B, Smallwood, A, Lai, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2.
Protein Sci., 17, 2008
|
|
5VT2
 
 | | Crystal structure of growth differentiation factor | | Descriptor: | 1,2-ETHANEDIOL, Growth/differentiation factor 15 | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2017-05-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Long-acting MIC-1/GDF15 molecules to treat obesity: Evidence from mice to monkeys.
Sci Transl Med, 9, 2017
|
|
1PSJ
 
 | | ACIDIC PHOSPHOLIPASE A2 FROM AGKISTRODON HALYS PALLAS | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Wang, X.Q, Lin, Z.J. | | Deposit date: | 1995-05-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acidic phospholipase A2 from the venom of Agkistrodon halys pallas at 2.0 A resolution.
J.Mol.Biol., 255, 1996
|
|
2N8A
 
 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
2MY1
 
 | | Solution structure of Bud31p | | Descriptor: | Pre-mRNA-splicing factor BUD31, ZINC ION | | Authors: | van Roon, A.M, Yang, J, Mathieu, D, Bermel, W, Nagai, K, Neuhaus, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | (113) Cd NMR Experiments Reveal an Unusual Metal Cluster in the Solution Structure of the Yeast Splicing Protein Bud31p.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5JYC
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EET hydrolysis intermediate | | Descriptor: | (5~{Z},11~{Z},14~{R},15~{R})-14,15-bis(oxidanyl)icosa-5,8,11-trienoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
