7DUN
 
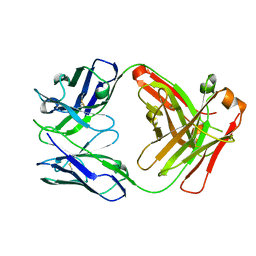 | |
7EXX
 
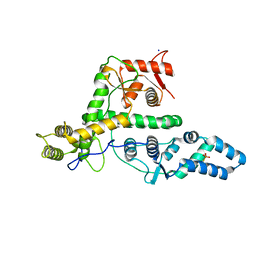 | | The structure of DndG | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptG, SODIUM ION | | Authors: | Wu, D, Wang, L. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
7CQI
 
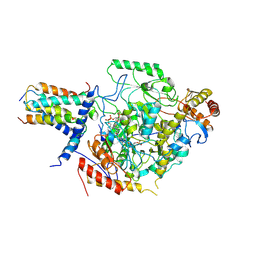 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CQK
 
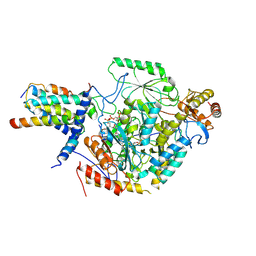 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7FJG
 
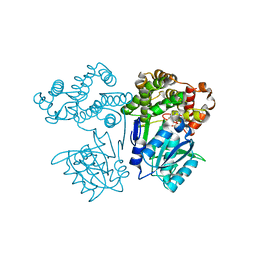 | | Crystal structure of butanol dehydrogenase A (YqdH) in complex with partial NADH from Fusobacterium nucleatum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, NADH-dependent butanol dehydrogenase A | | Authors: | Bai, X, Lan, J, Wang, L, Bu, T, Xu, Y. | | Deposit date: | 2021-08-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of butanol dehydrogenase A (YqdH) in complex with partial NADH from Fusobacterium nucleatum
To Be Published
|
|
7FDV
 
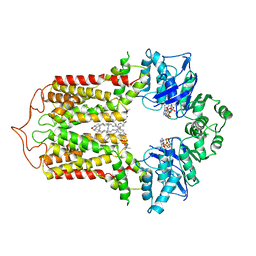 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | Authors: | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | Deposit date: | 2021-07-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
5J9S
 
 | | ENL YEATS in complex with histone H3 acetylation at K27 | | Descriptor: | H3K27ac peptide from Histone H3.1, Protein ENL, SULFATE ION | | Authors: | Li, Y, Li, H. | | Deposit date: | 2016-04-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Nature, 543, 2017
|
|
7F70
 
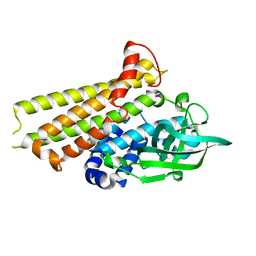 | | Crystal structure of Rv3094c | | Descriptor: | Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y, Zhang, H.T. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
7F74
 
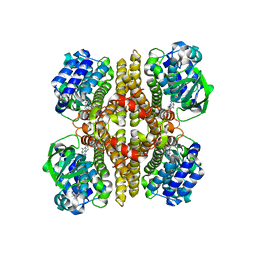 | | Rv3094c in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
7F72
 
 | | Rv3094c in complex with FAD and ETH. | | Descriptor: | 2-ethylpyridine-4-carboximidothioic acid, FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
5JMV
 
 | | Crystal structure of mjKae1-pfuPcc1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Probable bifunctional tRNA threonylcarbamoyladenosine biosynthesis protein, ... | | Authors: | Wan, L, Sicheri, F. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3864696 Å) | | Cite: | Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis.
Nucleic Acids Res., 44, 2016
|
|
7D0Z
 
 | |
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
7TB4
 
 | | Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Antibodies with potent and broad neutralizing activity against antigenically diverse and highly transmissible SARS-CoV-2 variants.
Biorxiv, 2021
|
|
8DZB
 
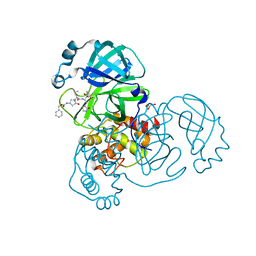 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 11 | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, benzyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-5-oxopyrrolidin-3-yl}carbamate | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
8DZC
 
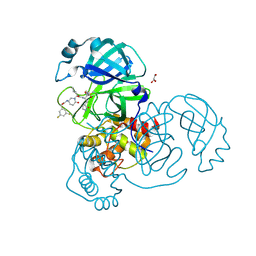 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 17 | | Descriptor: | (3,5-difluorophenyl)methyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-6-oxopiperidin-3-yl}carbamate, 3C-like proteinase nsp5, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
4U44
 
 | | MAP4K4 in complex with inhibitor (compound 16) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-phenyl-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U43
 
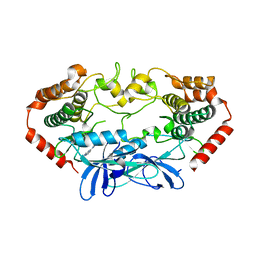 | | MAP4K4 in complex with inhibitor (compound 6) | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 4, N-(pyridin-3-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4U45
 
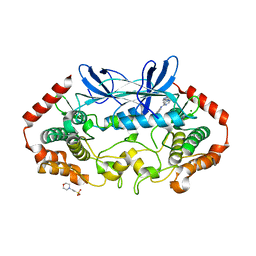 | | MAP4K4 in complex with inhibitor (compound 25) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(1H-pyrazol-4-yl)-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
8CEG
 
 | | BAR domain protein FAM92A1 essential for mitochondrial membrane remodeling | | Descriptor: | CBY1-interacting BAR domain-containing protein 1 | | Authors: | Kajander, T, Fudo, S, Yan, Z, Zhao, H. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Membrane remodeling by FAM92A1 during brain development regulates neuronal morphology, synaptic function, and cognition.
Nat Commun, 15, 2024
|
|
4ZPV
 
 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
4ZPW
 
 | | Structure of unbound MERS-CoV spike receptor-binding domain (England1 strain). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Spike glycoprotein | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
5UEW
 
 | | BRD2 Bromodomain2 with A-1360579 | | Descriptor: | Bromodomain-containing protein 2, N-[3-(4-methoxy-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UF0
 
 | | BRD4_BD2-A-35165 | | Descriptor: | 2-methyl-5-(methylamino)-6-phenylpyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
