2N3T
 
 | |
4DQO
 
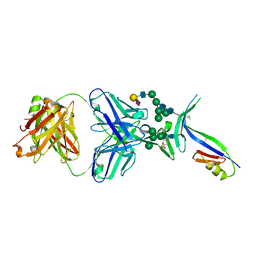 | | Crystal Structure of PG16 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | 1FD6-V1V2 scaffold ZM109 HIV-1 strain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 Fab Heavy Chain, ... | | Authors: | Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7MXD
 
 | |
3SSM
 
 | |
7N28
 
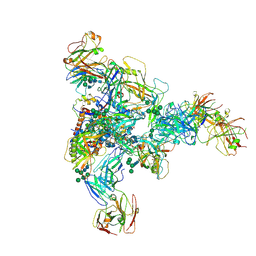 | |
7NB6
 
 | |
2N3W
 
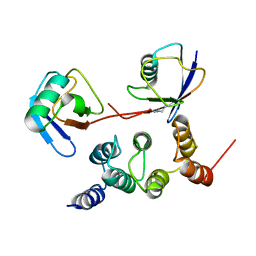 | |
4FCF
 
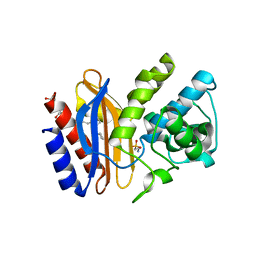 | | K234R: apo structure of inhibitor resistant beta-lactamase | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Rodkey, E.A, van den Akker, F. | | Deposit date: | 2012-05-24 | | Release date: | 2012-12-26 | | Last modified: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Design and exploration of novel boronic acid inhibitors reveals important interactions with a clavulanic acid-resistant sulfhydryl-variable (SHV) beta-lactamase.
J.Med.Chem., 56, 2013
|
|
7N9H
 
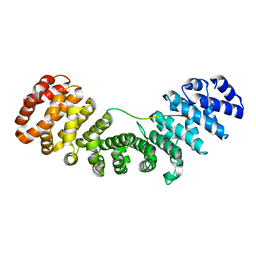 | |
2N9A
 
 | |
3UWP
 
 | | Crystal structure of Dot1l in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
3UQC
 
 | |
3V79
 
 | | Structure of human Notch1 transcription complex including CSL, RAM, ANK, and MAML-1 on HES-1 promoter DNA sequence | | Descriptor: | DNA 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', DNA 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Conformational Locking upon Cooperative Assembly of Notch Transcription Complexes.
Structure, 20, 2012
|
|
4DVR
 
 | |
3UXK
 
 | |
2OFO
 
 | | MSrecA-native | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2007-01-04 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRI
 
 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
2O9A
 
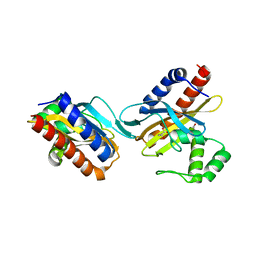 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
3T9C
 
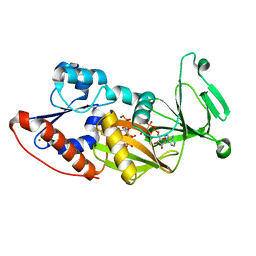 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and inositol hexakisphosphate (IP6) | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3TGS
 
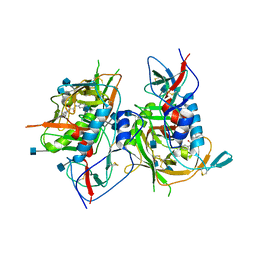 | | Crystal structure of HIV-1 clade C strain C1086 gp120 core in complex with NBD-556 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 core, N-(4-chlorophenyl)-N'-(2,2,6,6-tetramethylpiperidin-4-yl)ethanediamide | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGQ
 
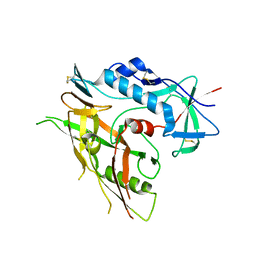 | | Crystal structure of unliganded HIV-1 clade B strain YU2 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 YU2 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2OAH
 
 | | Crystal Structure of Human Beta Secretase Complexed with inhibitor | | Descriptor: | Beta-secretase 1, N-[(1S,2S)-2-AMINO-1-(3-THIENYLMETHYL)HEXYL]-2-({[(1S,2S)-2-METHYLCYCLOPROPYL]METHYL}AMINO)-6-[METHYL(METHYLSULFONYL)AMINO]ISONICOTINAMIDE | | Authors: | Munshi, S. | | Deposit date: | 2006-12-15 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and SAR of isonicotinamide BACE-1 inhibitors that bind beta-secretase in a N-terminal 10s-loop down conformation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OEP
 
 | | MSrecA-ADP-complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
