3F4M
 
 | | Crystal structure of TIPE2 | | Descriptor: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | Authors: | Zhang, X, Wang, J, Shi, Y. | | Deposit date: | 2008-11-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
4DNW
 
 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
3P5N
 
 | |
4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
3H11
 
 | |
4F23
 
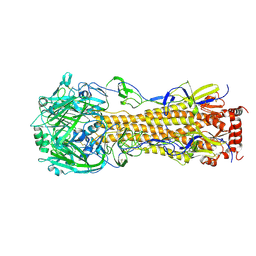 | | Influenza A virus hemagglutinin H16 HA0 structure with an alpha-helix conformation in the cleavage site: a potential drug target | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
3H4P
 
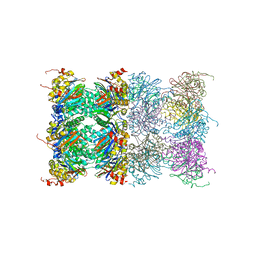 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H13
 
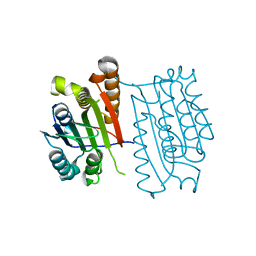 | | c-FLIPL protease-like domain | | Descriptor: | CASP8 and FADD-like apoptosis regulator | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of procaspase-8 activation by c-FLIPL.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H4M
 
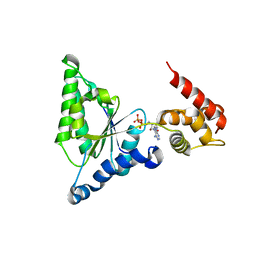 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H43
 
 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
7T0C
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2e | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-5-butyl-4-({1-[(4-chlorophenyl)methyl]-1H-imidazol-5-yl}methyl)-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T0D
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2k | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-propyl-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T0B
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2g | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-5-butyl-4-({1-[(4-fluorophenyl)methyl]-1H-imidazol-5-yl}methyl)-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T0E
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2b | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-(3-chlorophenyl)piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T0A
 
 | | Cryptococcus neoformans protein farnesyltransferase co-crystallized with FPP and inhibitor 2f | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (5S)-4-({1-[(4-bromophenyl)methyl]-1H-imidazol-5-yl}methyl)-5-butyl-1-[3-(trifluoromethoxy)phenyl]piperazin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T09
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2d | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, 4-{[5-({(2S)-2-butyl-5-oxo-4-[3-(trifluoromethoxy)phenyl]piperazin-1-yl}methyl)-1H-imidazol-1-yl]methyl}benzonitrile, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7T08
 
 | | Cryptococcus neoformans protein farnesyltransferase in complex with FPP and inhibitor 2q | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 1,2-ETHANEDIOL, 4-{[5-({(2S)-2-butyl-5-oxo-4-[3-(trifluoromethoxy)phenyl]piperazin-1-yl}methyl)-1H-imidazol-1-yl]methyl}-2-fluorobenzonitrile, ... | | Authors: | Wang, Y, Shi, Y, Beese, L.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure-Guided Discovery of Potent Antifungals that Prevent Ras Signaling by Inhibiting Protein Farnesyltransferase.
J.Med.Chem., 65, 2022
|
|
7TA8
 
 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
5IY3
 
 | | Zika Virus Non-structural Protein NS1 | | Descriptor: | Genome polyprotein | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JNX
 
 | | The 6.6 A cryo-EM structure of the full-length human NPC1 in complex with the cleaved glycoprotein of Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Wan, T, Shi, Y, Gao, F, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.56 Å) | | Cite: | Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection
Cell, 165, 2016
|
|
1KN5
 
 | | SOLUTION STRUCTURE OF ARID DOMAIN OF ADR6 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
5A63
 
 | | Cryo-EM structure of the human gamma-secretase complex at 3.4 angstrom resolution. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, X, Yan, C, Yang, G, Lu, P, Ma, D, Sun, L, Zhou, R, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An Atomic Structure of Human Gamma-Secretase
Nature, 525, 2015
|
|
1KKX
 
 | | Solution structure of the DNA-binding domain of ADR6 | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
8JOL
 
 | | cryo-EM structure of the CED-4/CED-3 holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 3, Cell death protein 4, ... | | Authors: | Li, Y, Tian, L, Zhang, Y, Shi, Y. | | Deposit date: | 2023-06-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
