2H7G
 
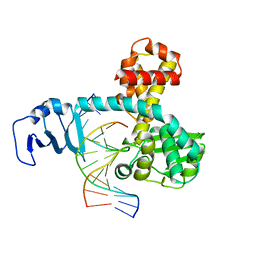 | | Structure of variola topoisomerase non-covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*TP*A)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
1XO0
 
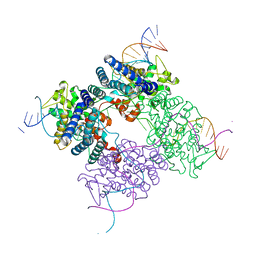 | | High resolution structure of the holliday junction intermediate in cre-loxp site-specific recombination | | Descriptor: | Recombinase CRE, loxP | | Authors: | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
1XNS
 
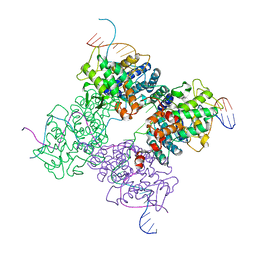 | | Peptide trapped Holliday junction intermediate in Cre-loxP recombination | | Descriptor: | Recombinase CRE, loxP DNA | | Authors: | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
1Y96
 
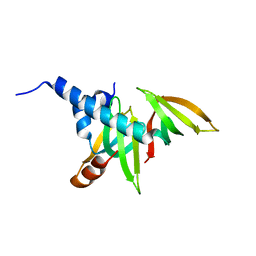 | | crystal structure of the Gemin6/Gemin7 heterodimer from the human SMN complex | | Descriptor: | Gem-associated protein 6, Gem-associated protein 7 | | Authors: | Ma, Y, Dostie, J, Dreyfuss, G, Van Duyne, G.D. | | Deposit date: | 2004-12-14 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The Gemin6-Gemin7 Heterodimer from the Survival of Motor Neurons Complex Has an Sm Protein-like Structure.
Structure, 13, 2005
|
|
1B4A
 
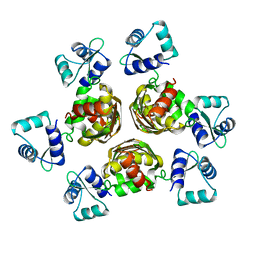 | | STRUCTURE OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
1B4B
 
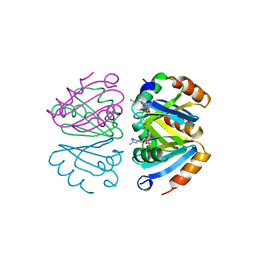 | | STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE, ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
1FKF
 
 | | ATOMIC STRUCTURE OF FKBP-FK506, AN IMMUNOPHILIN-IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Vanduyne, G.D, Standaert, R.F, Karplus, P.A, Schreiber, S.L, Clardy, J. | | Deposit date: | 1991-05-07 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex.
Science, 252, 1991
|
|
5J0N
 
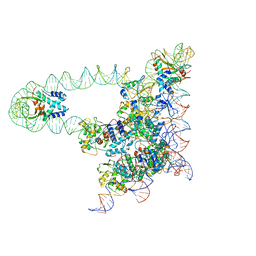 | | Lambda excision HJ intermediate | | Descriptor: | Excisionase, Integrase, Integration host factor subunit alpha, ... | | Authors: | Van Duyne, G, Grigorieff, N, Landy, A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structure of a Holliday junction complex reveals mechanisms governing a highly regulated DNA transaction.
Elife, 5, 2016
|
|
5JHQ
 
 | |
2CRX
 
 | |
6VRG
 
 | | Structure of HIV-1 integrase with native amino-terminal sequence | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Eilers, G, Gupta, K, Allen, A, Zhou, J, Hwang, Y, Cory, M, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2020-02-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influence of the amino-terminal sequence on the structure and function of HIV integrase.
Retrovirology, 17, 2020
|
|
8CT5
 
 | |
8CTA
 
 | | Minimal 2:2 Ternary Complex between BI-224436 bound HIV-1 Integrase Catalytic Core Domain Dimer and Carboxy Terminal Domains | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
8CT7
 
 | | Catalytic Core Domain of HIV-1 Integrase (F185K) bound with BI-224436 | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
3CRX
 
 | |
1FKK
 
 | | ATOMIC STRUCTURE OF FKBP12, AN IMMUNOPHILIN BINDING PROTEIN | | Descriptor: | FK506 BINDING PROTEIN, SULFATE ION | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1KBU
 
 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
2LEH
 
 | | Solution structure of the core SMN-Gemin2 complex | | Descriptor: | Survival motor neuron protein, Survival of motor neuron protein-interacting protein 1 | | Authors: | Sarachan, K.L, Valentine, K, Gupta, K, Moorman, V, Gledhill, J, Bernens, M, Tommos, C, Wand, A.J, Van Duyne, G. | | Deposit date: | 2011-06-15 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core SMN-Gemin2 complex.
Biochem.J., 445, 2012
|
|
1MA7
 
 | |
1D7H
 
 | | FKBP COMPLEXED WITH DMSO | | Descriptor: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7J
 
 | | FKBP COMPLEXED WITH 4-HYDROXY-2-BUTANONE | | Descriptor: | 4-HYDROXY-2-BUTANONE, AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7I
 
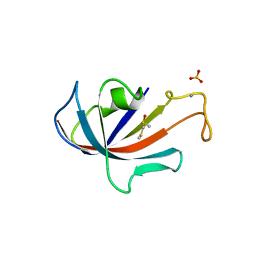 | | FKBP COMPLEXED WITH METHYL METHYLSULFINYLMETHYL SULFIDE (DSS) | | Descriptor: | AMMONIUM ION, METHYL METHYLSULFINYLMETHYL SULFIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1FKJ
 
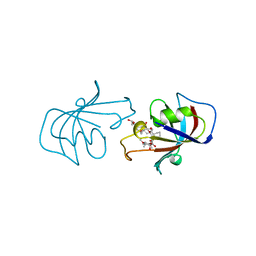 | | ATOMIC STRUCTURE OF FKBP12-FK506, AN IMMUNOPHILIN IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1FKL
 
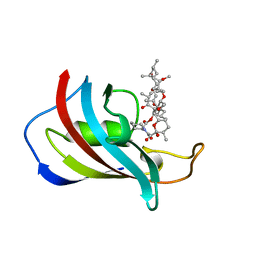 | | ATOMIC STRUCTURE OF FKBP12-RAPAYMYCIN, AN IMMUNOPHILIN-IMMUNOSUPPRESSANT COMPLEX | | Descriptor: | FK506 BINDING PROTEIN, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Wilson, K.P, Sintchak, M.D, Thomson, J.A, Navia, M.A. | | Deposit date: | 1995-08-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparative X-ray structures of the major binding protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
4OJR
 
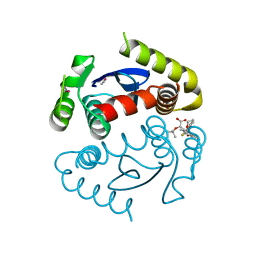 | | Crystal Structure of the HIV-1 Integrase catalytic domain with GSK1264 | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, CACODYLATE ION, HIV-1 Integrase, ... | | Authors: | Gupta, K, Brady, T, Dyer, B, Hwang, Y, Male, F, Nolte, R.T, Wang, L, Velthuisen, E, Jeffrey, J, Van Duyne, G, Bushman, F.D. | | Deposit date: | 2014-01-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Allosteric Inhibition of Human Immunodeficiency Virus Integrase: LATE BLOCK DURING VIRAL REPLICATION AND ABNORMAL MULTIMERIZATION INVOLVING SPECIFIC PROTEIN DOMAINS.
J.Biol.Chem., 289, 2014
|
|
