8EHO
 
 | | PRRSV-1 PLP2 domain bound to ubiquitin | | Descriptor: | 3-AMINOPROPANE, GLYCEROL, NITRATE ION, ... | | Authors: | Bailey-Elkin, B.A, Mark, B.L. | | Deposit date: | 2022-09-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Demonstrating the importance of porcine reproductive and respiratory syndrome virus papain-like protease 2 deubiquitinating activity in viral replication by structure-guided mutagenesis.
Plos Pathog., 19, 2023
|
|
8EHN
 
 | | PRRSV-1 PLP2 domain | | Descriptor: | ACETATE ION, Papain-like protease 2, ZINC ION | | Authors: | Bailey-Elkin, B.A, Mark, B.L. | | Deposit date: | 2022-09-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Demonstrating the importance of porcine reproductive and respiratory syndrome virus papain-like protease 2 deubiquitinating activity in viral replication by structure-guided mutagenesis.
Plos Pathog., 19, 2023
|
|
3QBX
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to 1,6-anhydro-n-actetylmuramic acid | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
3QBW
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
3PSE
 
 | | Structure of a viral OTU domain protease bound to interferon-stimulated gene 15 (ISG15) | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, GLYCEROL, RNA polymerase, ... | | Authors: | Bacik, J.P, James, T.W, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-01 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PT2
 
 | | Structure of a viral OTU domain protease bound to Ubiquitin | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, ACETATE ION, RNA polymerase, ... | | Authors: | James, T.W, Bacik, J.P, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8CX9
 
 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | Descriptor: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | Deposit date: | 2022-05-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
4MO4
 
 | | Crystal structure of AnmK bound to AMPPCP | | Descriptor: | Anhydro-N-acetylmuramic acid kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformational Itinerary of Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase during Its Catalytic Cycle.
J.Biol.Chem., 289, 2014
|
|
4RF1
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
5V6A
 
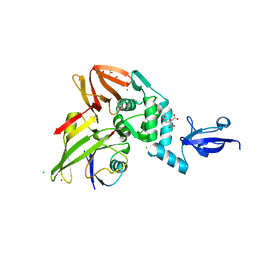 | |
5V5I
 
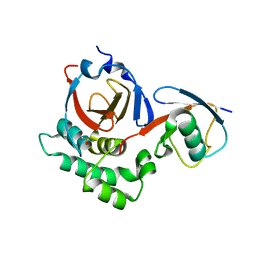 | |
5VQE
 
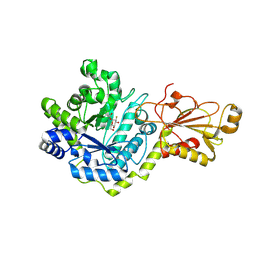 | | Beta-glucoside phosphorylase BglX bound to 2FGlc | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucoside phosphorylase BglX | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
5UTP
 
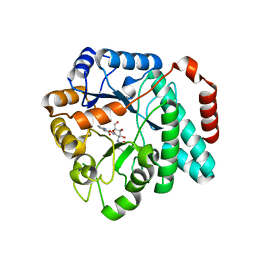 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to N-ethylbutyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, N-[(2Z,3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-2-{[(phenylcarbamoyl)oxy]imino}tetrahydro-2H-pyran-3-yl]-2-ethylbutanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
5V5H
 
 | |
5V69
 
 | |
5UTQ
 
 | |
5V5G
 
 | |
5UTR
 
 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-butyryl-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]butanamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2017-02-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility of the glycosidase NagZ allows it to bind structurally diverse inhibitors to suppress beta-lactam antibiotic resistance.
Protein Sci., 26, 2017
|
|
5VQD
 
 | | Beta-glucoside phosphorylase BglX | | Descriptor: | Beta-glucoside phosphorylase BglX, GLYCEROL | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
4GVH
 
 | |
4GYJ
 
 | |
4GVG
 
 | |
4GVF
 
 | |
4GYK
 
 | |
4GVI
 
 | | Crystal structure of mutant (D248N) Salmonella typhimurium family 3 glycoside hydrolase (NagZ) in complex with GlcNAc-1,6-anhMurNAc | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bacik, J.P, Mark, B.L. | | Deposit date: | 2012-08-30 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active Site Plasticity within the Glycoside Hydrolase NagZ Underlies a Dynamic Mechanism of Substrate Distortion.
Chem.Biol., 19, 2012
|
|
