4TU6
 
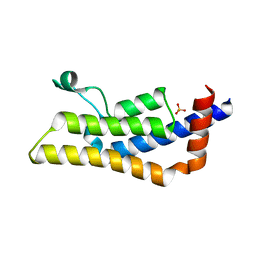 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU4
 
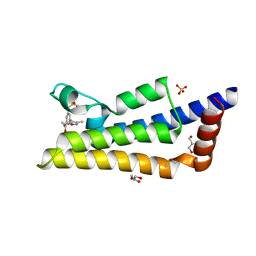 | | Crystal structure of ATAD2A bromodomain complexed with 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoicacid | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoic acid, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT6
 
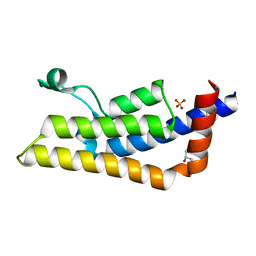 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
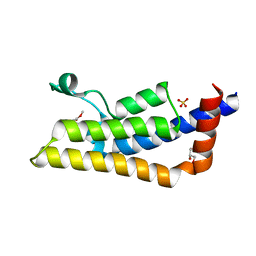 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
7LUB
 
 | |
4NY0
 
 | |
7B1O
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complex with compound 22 | | Descriptor: | 4-chloranyl-N-[(1R)-1-[(1S,5R)-3-quinolin-4-yloxy-6-bicyclo[3.1.0]hexanyl]propyl]benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lammens, A, Krapp, S, Lewis, R.T, Hamilton, M.M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
2KW6
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 1 (CDK2-associated protein 1; oral cancer suppressor Deleted in oral cancer 1, DOC-1) from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3057H | | Descriptor: | Cyclin-dependent kinase 2-associated protein 1 | | Authors: | Ertekin, A, Aramini, J.M, Rossi, P, Lee, A.B, Jiang, M, Ciccosanti, C.T, Xiao, R, Swapna, G.V.T, Rost, B, Everett, J.K, Acton, T.B, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Human cyclin-dependent kinase 2-associated protein 1 (CDK2AP1) is dimeric in its disulfide-reduced state, with natively disordered N-terminal region.
J.Biol.Chem., 287, 2012
|
|
4WUH
 
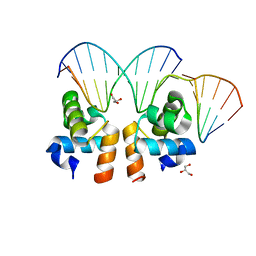 | | Crystal structure of E. faecalis DNA binding domain LiaR wild type complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
4WSZ
 
 | |
4WT0
 
 | |
4WUL
 
 | |
4WU4
 
 | | Crystal structure of E. faecalis DNA binding domain LiaRD191N complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*GP*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), GLYCEROL, ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
5HEV
 
 | |
3R66
 
 | | Crystal structure of human ISG15 in complex with NS1 N-terminal region from influenza virus B, Northeast Structural Genomics Consortium Target IDs HX6481, HR2873, and OR2 | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Guan, R, Ma, L.C, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the complex of ISG15 and Influenza B virus NS1 Protein: implications for the mechanism of SN1-mediated inhibition of ISG15 conjugation
To be Published
|
|
3SDL
 
 | | Crystal structure of human ISG15 in complex with NS1 N-terminal region from influenza B virus, Northeast Structural Genomics Consortium Target IDs HX6481, HR2873, and OR2 | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Guan, R, Ma, L.-C, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
