6NDO
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant L193N from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Rinaldi, J, Conforte, V, Malamud, F, Goldbaum, F.A, Chavas, L, Bonomi, H.R. | | Deposit date: | 2018-12-14 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
6D3G
 
 | | PER-2 class A extended-spectrum beta-lactamase crystal structure in complex with avibactam at 2.4 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, TETRAETHYLENE GLYCOL | | Authors: | Power, P, Ruggiero, M, Gutkind, G, Bonomo, R, Klinke, S. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural Insights into the Inhibition of the Extended-Spectrum beta-Lactamase PER-2 by Avibactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6DGU
 
 | | PER-2 class A extended-spectrum beta-lactamase crystal structure at 2.69 Angstrom resolution | | Descriptor: | Beta-lactamase | | Authors: | Power, P, Ruggiero, M, Gutkind, G, Bonomo, R, Klinke, S. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Structural Insights into the Inhibition of the Extended-Spectrum beta-Lactamase PER-2 by Avibactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
7LRH
 
 | | C-terminal domain of RibD from Brucella abortus (5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5'-phosphate reductase) | | Descriptor: | Riboflavin biosynthesis protein RibD, SULFATE ION | | Authors: | Bonomi, H.R, Cerutti, M.L, Posadas, D.M, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2021-02-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-terminal domain of RibD from Brucella abortus (5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5'-phosphate reductase)
To be published
|
|
7MHW
 
 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
2QT7
 
 | | Crystallographic structure of the mature ectodomain of the human receptor-type protein-tyrosine phosphatase IA-2 at 1.30 Angstroms | | Descriptor: | CALCIUM ION, Receptor-type tyrosine-protein phosphatase-like N | | Authors: | Primo, M.E, Klinke, S, Sica, M.P, Goldbaum, F.A, Jakoncic, J, Poskus, E, Ermacora, M.R. | | Deposit date: | 2007-08-01 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the Mature Ectodomain of the Human Receptor-type Protein-tyrosine Phosphatase IA-2
J.Biol.Chem., 283, 2008
|
|
2Q76
 
 | | Mouse anti-hen egg white lysozyme antibody F10.6.6 Fab fragment | | Descriptor: | Fab F10.6.6 fragment Heavy Chain, Fab F10.6.6 fragment Light Chain | | Authors: | Cauerhff, A, Klinke, S, Acierno, J.P, Goldbaum, F.A, Braden, B.C. | | Deposit date: | 2007-06-06 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity maturation increases the stability and plasticity of the Fv domain of anti-protein antibodies.
J.Mol.Biol., 374, 2007
|
|
5AKP
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris bound to BV chromophore | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BILIVERDINE IX ALPHA, CHLORIDE ION, ... | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2015-03-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas Campestris Provides Clues to its Long-Range Signaling Mechanism.
J.Mol.Biol., 428, 2016
|
|
5ANP
 
 | |
7L5A
 
 | | Crystal structure of the photosensory module from Xanthomonas campestris bacteriophytochrome XccBphP in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7L59
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP-G454E variant from Xanthomonas campestris in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
6OVP
 
 | |
6ODD
 
 | | Crystal structure of the human complex ACP-ISD11 | | Descriptor: | Acyl carrier protein, mitochondrial, CALCIUM ION, ... | | Authors: | Herrera, M.G, Noguera, M.E, Klinke, S, Santos, J. | | Deposit date: | 2019-03-26 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Human ACP-ISD11 Heterodimer.
Biochemistry, 58, 2019
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7KVC
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
4D6Y
 
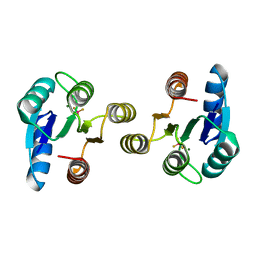 | | Crystal structure of the receiver domain of NtrX from Brucella abortus in complex with beryllofluoride and magnesium | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Otero, L.H, Fernandez, I, Carrica, M.C, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
6BN3
 
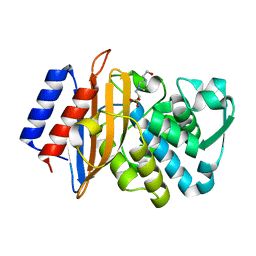 | | CTX-M-151 class A extended-spectrum beta-lactamase apo crystal structure at 1.3 Angstrom resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Rodriguez, M.M, Gutkind, G, Ishii, Y, Bonomo, R.A, Klinke, S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Structural and Biochemical Characterization of the Novel CTX-M-151 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam.
Antimicrob.Agents Chemother., 65, 2021
|
|
6BPF
 
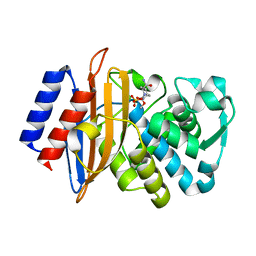 | | CTX-M-151 class A extended-spectrum beta-lactamase crystal structure in complex with avibactam at 1.32 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Rodriguez, M.M, Gutkind, G, Ishii, Y, Bonomo, R.A, Klinke, S. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Structural and Biochemical Characterization of the Novel CTX-M-151 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam.
Antimicrob.Agents Chemother., 65, 2021
|
|
6BNX
 
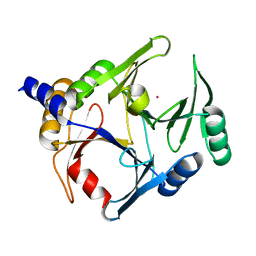 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P6(3) | | Descriptor: | COBALT (II) ION, Lactoylglutathione lyase | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
6BNZ
 
 | | Crystal structure of E144Q-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
6BNN
 
 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
4E0F
 
 | | Crystallographic structure of trimeric Riboflavin Synthase from Brucella abortus in complex with riboflavin | | Descriptor: | RIBOFLAVIN, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-03-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4FXU
 
 | | Crystallographic structure of trimeric riboflavin synthase from Brucella abortus | | Descriptor: | Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-07-03 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4G6I
 
 | | Crystallographic structure of trimeric riboflavin synthase from Brucella abortus in complex with roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-07-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8DQT
 
 | | Human PDK1 kinase domain in complex with Valsartan | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL | | Authors: | Gross, L.Z.F, Klinke, S, Biondi, R.M. | | Deposit date: | 2022-07-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein.
Sci.Signal., 16, 2023
|
|
